Metatranscriptomic holobiont analysis of carbohydrate-active enzymes in the millipede Telodeinopus aoutii (Diplopoda, Spirostreptida)
- 1Biology Centre AS CR, Institute of Soil Biology, České Budějovice, Czechia
- 2Faculty of Agriculture and Technology, University of South Bohemia in České Budějovice, České Budějovice, Czechia
As important decomposers of soil organic matter, millipedes contribute to lignocellulose decomposition and nutrient cycling. The degradation of lignocellulose requires the action of several carbohydrate-active enzymes (CAZymes) and, in most invertebrates, depends on the activity of mutualistic gut microorganisms. To address the question of the importance of the microbiota and endogenous (host) enzymes in digestive processes in millipedes, we analyzed metatranscriptomic data from the tropical millipede Telodeinopus aoutii at the holobiont level. Functional annotation included identification of expressed CAZymes (CAZy families and EC terms) in the host and its intestinal microbiota, foregut, midgut, and hindgut, compared to non-intestinal tissues. Most of the 175 CAZy families were expressed exclusively in the gut microbiota and more than 50% of these microbial families were expressed exclusively in the hindgut. The greatest diversity of expressed endogenous CAZymes from all gut sections was found in the midgut (77 families). Bacteria were the major microbial producers of CAZymes, Proteobacteria dominating in the midgut and Bacteriodetes with Firmicutes in the hindgut. The contribution of the eukaryotic microbiota to CAZymes production was negligible. Functional classification of expressed CAZy families confirmed a broad functional spectrum of CAZymes potentially expressed in the holobiont. Degradation of lignocellulose in the digestive tract of the millipede T. aoutii depends largely on bacterial enzymes expressed in the hindgut. Endogenous cellulases were not detected, except for the potentially cellulolytic family AA15, but an expression of cellulolytic enzymes of this family was not confirmed at the EC-number level. The midgut had the greatest diversity of expressed endogenous CAZymes, mainly amylases, indicating the importance of digesting α-glucosidases for the millipede. In contrast, bacterial lignocellulolytic enzymes are sparsely expressed here. The hindgut was the hotspot of microbial degradation of cellulose and hemicellulases. The gain of the millipede from the microbial lignocellulose degradation in the gut, and consequently the mutualistic status of the relationship between the millipede and its cellulolytic gut bacteria, depends on the ability of the millipede to take up microbial metabolites as nutrients through the hindgut wall. Enzymes expressed in the intestine can degrade all components of lignocellulose except lignin. Assuming that soil microbiota is partially degraded lignin in the millipede diet, T. aoutii can be considered a decomposer of soil organic matter relying primarily on its gut bacteria. The deposition of millipede fecal pellets containing an organic matter modified by the hindgut bacterial community could be of ecological significance.
Introduction
Saprophagous soil macrofauna plays an important role in the transformation of leaf litter. Soil invertebrates feed on decaying plant material colonized by microbial decomposers (bacteria and fungi) (David, 2015), mechanically and chemically alter the consumed soil organic matter, and produce a variety of excrements that support soil formation. The dead plant organic matter consists of a complex of cellulose, hemicellulose, and lignin. To digest and utilize it, the synergistic action of different enzymes is required (Ezeilo et al., 2017). The degradation of the lignocellulosic complex is mainly the domain of cellulolytic protists, bacteria, and fungi. Xylophagous arthropods have supplemented the nutritional potential of their diet by forming relationships with protozoa, fungi, and bacteria (Kane, 1997). Despite the growing list of newly discovered endogenous cellulases in animals (Watanabe and Tokuda, 2001), a complex synergistic action of endogenous digestive enzymes from arthropods and gut microbial activity plays a role in the degradation of lignocellulose in the intestine of some invertebrates (termites, beetles, woodlice). In specialized xylophagous insects (such as some termite groups) with high assimilation efficiency of cellulose, the gut anatomy is adapted to harbor cellulolytic microbial symbionts. The role of these symbionts is critical and well described (Kane, 1997). For macrofauna feeding on litter, such as millipedes, the importance of cellulose, hemicellulose, and lignin utilization and the precise role of gut microorganisms are not clearly understood. There is some evidence, that invertebrates chemically degrade dead plant material and may play a larger role in terrestrial degradation pathways than previously thought (Griffiths et al., 2021). Scharf, 2015 promoted the idea of the digestome, which describes the combined enzymes produced by microbes and invertebrates that interact in plant decomposition. The assumption that invertebrates were not directly involved in the decomposition of dead plant material was based on the notion that animal decomposers did not produce endogenous cellulases and have low assimilation efficiencies (Van der Drift, 1951). Millipedes consumed food with a high content (up to 50%) of lignocellulose, which was usually partially processed by free-living soil microorganisms. Their assimilation efficiency of about 30% (Hopkin and Read, 1992) suggests that the energy contained in the cellulose may not be utilized. According to a more recent review (David, 2014), laboratory estimates of assimilation efficiencies of macroarthropods vary considerably (from less than 5% to more than 50%) in different studies.
Energy budget experiments showed that the metabolic rate of millipedes fed pure cellulose was as low as starved individuals, suggesting that millipedes fed pure cellulose cannot maintain a positive energy balance without the addition of a nitrogen source (Šustr et al., 2020b). However, older experiments on some millipede species with isotopically labeled cellulose appear to confirm the utilization of carbon in cellulose (Taylor, 1982; Bignell, 1989). In vitro, a wide range of enzyme activities, including cellulases, has been detected in crude millipede intestinal homogenates using enzymatic assays (Šustr, 1999; Šustr et al., 2020a). However, this type of measurement did not provide information on the origin of the enzymes and the actual rate of substrate degradation in vivo. The intestinal tract of millipedes is a straight tube without distinct regions specialized for the harboring of intestinal symbionts (Hopkin and Read, 1992). However, anoxia and reducing conditions in the gut contents (Horváthová et al., 2021) and the structured inner surface of the hindgut wall (Fontanetti et al., 2015) promote the proliferation of the fermenting microbial community. Their activity in the gut has been demonstrated by the presence of fermentation end-products (short-chain fatty acids and hydrogen) and methane production in the millipedes Archispirostreptus gigas (Spirostreptida) and Epibolus pulchripes (Spirobolida) (Horváthová et al., 2021).
The microbial community in the millipede intestine is rich and diverse (Byzov, 2006), including potentially cellulolytic groups (Taylor, 1982). However, the stable symbiotic microbiota has not been confirmed, and the proportion of transient free-living microorganisms ingested with food has not yet been clearly described. Recent studies, using genome or transcriptome sequencing and proteome or metabolome analysis, have ushered in a new era to elucidate the role of plant cell wall-degrading enzymes in the digestive physiology of insects (Tokuda, 2019) and other invertebrates (Bredon et al., 2018). Recently expanded database information, including functional aspects such as information on glycoside hydrolases (GH), supports omics studies. GH, according to the International Union of Biochemistry and Molecular Biology enzyme nomenclature (EC 3.2.1.-), represent a widespread group of enzymes that hydrolyze the glycosidic bond between carbohydrates or between a carbohydrate and a non-carbohydrate moiety. The EC nomenclature of GHs is based on their substrate specificity and occasionally on their molecular mechanism. To reflect structural features and evolutionary relationships among these enzymes, a classification of GHs into families based on amino acid sequence similarities has been proposed. The Carbohydrate-Active Enzymes database (CAZy) (Drula et al., 2022) provides a continuously updated list of carbohydrate-active enzymes (CAZymes) in general. CAZymes are classified into protein families associated with characterized module classes of modules. In addition to GH, these include glycosyltransferases (GT), polysaccharide lyases (PL), carbohydrate esterases (CE), auxiliary activities (AA), and carbohydrate-binding modules (CBM). These enzymes are involved in the synthesis and breakdown of glycoconjugates, oligo- and polysaccharides (Lombard et al., 2014). Of the ecologically important lignocellulolytic enzymes, most cellulases belong to the GHs, many hemicellulases to the CEs, and GHs. Lignin modifying enzymes (LME) are often classified as AAs, breaking down, unlike GHs, lignocellulolytic matter with oxidative processes (Bredon et al., 2020). As the CAZy database administrators pointed out, the modularity of these enzymes and the grouping of enzymes with different substrate specificity in the same sequence-based families can complicate the functional annotation of CAZymes. Nevertheless, the CAZy database represents a very useful tool widely used by the scientific community, mostly in combination with the recognition of enzyme activities described by EC numbers in omics data (Bredon et al., 2018, 2020; Ceja-Navarro et al., 2019; for example).
The aim of this study was to evaluate the role of the host and microbiota in enzymatic polysaccharide degradation in the millipede holobiont using functional annotation of the metatranscriptome. The large tropical millipede species Telodeinopus aoutii was used as a model species for analysis of gene expression (host and microbial) in the gut (foregut, midgut, and hindgut) and non-intestinal tissues. The expression of proteins belonging to CAZy families and Enzyme Commission terms (EC) corresponding to the activities of carbohydrase enzymes was analyzed. The distribution of the expression among the holobiont parts (intestinal foregut, midgut and hindgut, and non-intestinal millipede tissues) and among the major groups of organisms in the holobiont (i.e., the millipede, ciliates, nematodes, fungi), bacteria and archaea were compared.
Materials and methods
Experimental animals
Specimens of T. aoutii (Demange, 1971) (Diplopoda, Spirostreptida, Spirostreptidae) were obtained from pet shops in the Czech Republic and kept under laboratory conditions. The millipedes were kept at the Institute of Soil Biology in plastic boxes (60 × 30 × 20 cm) at 25°C on the forest floor substrate containing peat, rotten wood, and a mixture of leaf litter (maple, oak, and beech). The lids of the boxes were ventilated; the substrate was regularly moistened. The leaf litter formed the main millipede diet. Vegetable pieces served as an additional food source. Calcium was added to the substrate in the form of sepia powder and aquarium fish food served as a nitrogen source. Coprophagy of millipede fecal pellets was not prevented. Only animals with full digestive tracts were used for gut dissection and subsequent analyses.
Holobiont metatranscriptomics
Four millipedes were cleaned with RNase AWAY™ (Sigma-Aldrich®) and sacrificed by freezing at –80°C for 10 min. Total RNA was extracted from the intestinal compartments [foregut (FG), midgut (MG), and hindgut (HG)], and from the remainder of the body (NG) using the RNeasy Power Soil Total RNA Kit, QIAGEN®. Salivary glands were analyzed together with FG. Crude RNA was precipitated with a glycogen-ethanol-acetate solution and resuspended in DEPC water (Top-Bio, Prague, Czechia). DNA was digested using the TURBO DNA-freeTM Kit (ThermoFisher Scientific®), and RNA was purified using the AllPrep DNA/RNA Micro Kit (QIAGEN®). The efficiency of DNA digestion was verified by the amplification of 16S rRNA genes. RNA integrity was checked by RNA gel electrophoresis and quantified on an RNA Nano Chip (Agilent® 2100 Bioanalyzer). All RNA samples were stored at –80°C until shipment to the sequencing laboratory at the UIC’s Sequencing Core, Chicago, IL, United States. Briefly, RNA-Seq libraries were prepared from approximately 120 ng of total RNA. The rRNA removal method followed a previously described methodology (Bogdanova et al., 2011) with a modification that enabled simultaneous depletion of host and microbial rRNA. Libraries were prepared using the Zymo-Seq RiboFree Total RNA Library Kit from ZYMO RESEARCH® according to the manufacturer’s instructions. RNA-Seq libraries were sequenced on an Illumina platform (2 × 100 bp paired-end Illumina MiSeq). Sequence files were deposited in the NCBI Sequence Read Archive (BioProject ID PRJNA749320).
All sequences were processed locally on computers running Linux Ubuntu. SortMeRNA (v2.1) (Kopylova et al., 2012) was used to remove rRNA reads from the datasets using the following databases: SILVA 16S and 23S for archaeal and bacterial rRNA, SILVA 18S and 28S for eukaryotic rRNA, and Rfam for 5S and 5.8S. Because the reference genomes and transcriptomes of T. aoutii are not available in public databases, transcripts were assembled de novo using the Trinity (v2.10.0) (Grabherr et al., 2011).
We reconstructed the transcripts for each millipede body segment (FG, MG, HG, and NG) using four biological replicates. Quality trimming and in silico normalization of reads were performed with default parameters in Trinity using Trimmomatic (Bolger et al., 2014) and the Trinity script insilico_read_normalization.pl, respectively. Initial assembly statistics were computed using the TrinityStats.pl script within the Trinity package. Transcripts were clustered using the CD-Hit package (Li and Godzik, 2006) with an identity threshold of 95%. A mapping-based method using Salmon (Patro et al., 2017) was used to quantify transcript abundance. Reads from each biological replicate were mapped against the assembly of the corresponding holobiont part (i.e., either from FG, MG, HG, or NG). The transcript quantification procedure yielded normalized expression values of transcripts in the sample, measured as transcripts per million (TPM) (Wagner et al., 2012).
Carbohydrate-active enzyme annotation from host and gut-biota
All open reading frames (ORFs) were predicted from metatranscriptomes using Transdecoder (v5.5.0) (Haas et al., 2013) with default parameters. ORFs were compared with the Non-Redundant protein database (July 1, 2020) using BLASTp (Altschul et al., 1990) with DIAMOND (Buchfink et al., 2015) in “–more-sensitive” mode with an E-value cut-off of 1e-5. The BLAST outputs were then imported into MEGAN6 (v6.19.5) (Huson et al., 2016) for segregating ORFs derived from host and microbiota. Each ORF was thus assigned to the most accurate taxonomic rank (i.e., kingdom, phylum, class, order, family, genus, and species) based on the LCA (lowest common ancestor) algorithm. The command-line version of eggNOG mapper v2 (Huerta-Cepas et al., 2017) was used to assign general functional annotation using precomputed orthologous groups from the eggNOG database (Huerta-Cepas et al., 2019).
For more specific functionality, CAZymes were identified using the Carbohydrate Active Enzyme (CAZy) database (Lombard et al., 2014). dbCAN2 (Zhang et al., 2018) was used to identify CAZy families, i.e., GHs, glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), auxiliary activities (AAs), and carbohydrate-binding modules (CBMs) from the previously filtered ORFs from the metatranscriptomes. dbCAN2 integrates three tools for annotating CAZymes: (1) HMMER (version 3.2.1) (Mistry et al., 2013) which uses the dbCAN CAZyme domain HMM database (Yin et al., 2012) for domain predictions, (2) DIAMOND for sequence comparisons against a custom pre-annotated CAZyme sequence database, and (3) HOTPEP (Busk et al., 2017), which performs searches against a conserved CAZyme short peptide database. dbCAN2 was run in a conservative manner, only CAZyme predicted by all three of the above tools was retained for subsequent analyses. As recommended by the dbCAN2 authors, CAZyme counts resulting from dbCAN assignment using the HMMER tool were considered for subsequent analyses.
Carbohydrate active enzyme families known to potentially contribute to lignocellulose degradation were selected for further analysis. Each individual CAZy family may represent enzymes involved in a variety of carbohydrate-modifying activities, including the degradation of lignocellulose. Therefore, the enzymatic activities of CAZymes were predicted using Hotpep to confirm their involvement in carbohydrate degradation and assign a corresponding Enzyme Commission number (EC). When Hotpep failed to predict the CAZyme function, the most common activity was retrieved manually from the CAZy database. Note that “CAZyme” refers to functional modules or domains, not the transcript itself. A single transcript may contain one, several, or no CAZy module. Therefore, the expression of CAZymes in terms of TPM values is only meaningful up to the CAZy family level, but not for the individual CAZy modules and their corresponding EC numbers. Therefore, as mentioned above, the module numbers predicted by HMMER- for CAZyme were used for most analyses in our study instead of TPM.
Substrate classification of expressed enzyme commission numbers and carbohydrate-active enzyme families from literature review
The EC numbers for the enzymes that degrade trehalose, starch, sucrose, hemicellulose, cellulose, laminarin, lichenin, chitin, pectin, and lignin were selected from a literature review (Supplementary Table S1). The expression of the enzymes from this list was checked to show the range of saccharolytic activities in the holobiont. The substrate specificities of the expressed CAZy families were classified by comparing the list in Supplementary Table S1 with activities documented in the database1 (November 2021).
Visualization and graphical representation
Venn diagrams (Venn, 1880) with different CAZyme counts were generated using the library (“VennDiagram”) in R (R Core Team., 2016). Counts of individual CAZy modules in each CAZyme-producing taxa were imported into Cytoscape 3.8.2 to build the interconnection and association map. Heat maps for the individual CAZyme families were generated using TPM expression matrices with Hclust22. Hierarchical clustering of sample types (i.e., taxonomic origin and tissue origin) and CAZy modules were performed using the Bray-Curtis dissimilarity. Circos plots (Krzywinski et al., 2009) were generated using Circos Table Viewer v0.63-93). To display the metabolic maps shown in the supplements, the KEGG pathway map 00500 (Starch and sucrose metabolism in the KEGG database, Kanehisa and Goto, 2000) was overlaid with omics data.
Results and discussion
Identification and classification of carbohydrate-active enzyme in the holobiont
The holobiont metatranscriptome assemblies of T. aoutii were analyzed for CAZymes encoding genes. The dbCAN pipeline identified a total of 1,337 CAZy modules in the host (Figure 1A) and 806 CAZy modules in the microbiome (Figure 1B). The CAZyme-associated genes containing the CAZy modules were further classified into enzyme families according to the CAZy nomenclature (Lombard et al., 2014). A total of 172 CAZy families were identified in the T. aoutii holobiont, including all known CAZy classes (i.e., AAs, CBMs, CEs, GHs, GTs, and PLs). GTs were the most abundant class in the T. aoutii holobiont with 49 different families, corresponding to 498 and 131 modules in the host and microbiota, respectively. Among the major CAZy classes, GHs and CBMs were frequently expressed in both, the microbiota, and the host. The higher proportion of expressed GTs is typical for the host, the expression of polysaccharide lyases, and a higher proportion of auxiliary activities for the microbial community. Glycosyltransferases were found in both eukaryotes and prokaryotes and catalyze the formation of the glycosidic bond to form glycoside, but they are not involved in the digestion process. Glycosylation is the common cellular modification of proteins and lipids; that is important for multicellular life (Kellokumpu et al., 2016). This explains the relatively high expression diversity of GTs in the host in the holobiont of T. aoutii. The second most important class of CAZymes in the T. aoutii holobiont was GH, with 343 and 289 modules in the host and microbiota, respectively, distributed among 63 different families. Of the 172 CAZy families identified in the T. aoutii holobiont, 46 and 94 families were specific to the host and its microbiota, respectively, and 35 were present in both (Figure 1C). This suggests that the gut microbiota plays an important role in millipedes, as previously shown in higher termites (Brune and Ohkuma, 2011).
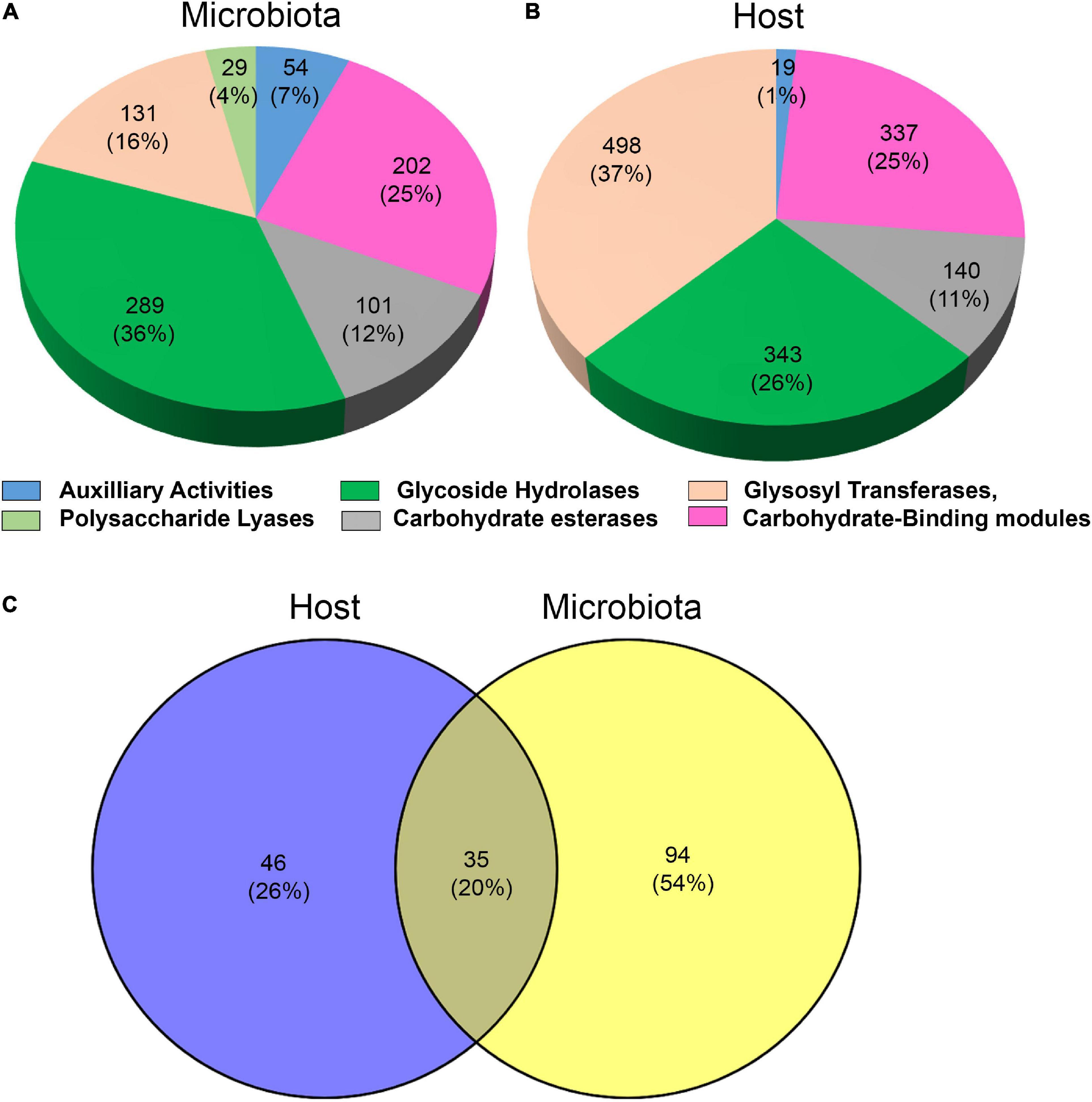
Figure 1. Major CAZy classes are present in the microbiota (A) and in the host (B). Total number of CAZy families found in the holobiont and their shares in the host and microbiota (C).
The GH13 family, which is known to include enzyme activities such as amylases, α-glucosidase, and starch debranching enzymes, was the most highly expressed GHs in the T. aoutii holobiont with 95 CAZy modules (Supplementary Figure S1). It was expressed in the host (73 modules) and in the microbiome (22 modules). Carbohydrate-binding modules (CBMs) in the T. aoutii holobiont represented 43 different families, corresponding to 497 and 202 modules in the host and microbiome, respectively. Among this class of modules, the CBM14 family, a chitin-binding module, was the most prominent in the holobiont of T. aoutii. It was expressed in the host (Figure 2), with 242 modules, accounting for 12.3% of the total host CAZy modules. Another abundant CBM family was CBM32, including galactose- and lactose binding-modules present in both the host and microbiome. In contrast, the CBM37 family, a family with broad binding specificity for xylan, chitin, microcrystalline, and phosphoric-acid swollen cellulose, was produced only by the microbiome (17 modules) in MG and HG, representing 2.1% of the total CAZy modules in the microbiome. Twelve families of CEs were represented by 215 and 101 modules in the host and the microbiome, respectively. Among the CEs, the CE1 and CE10 families were the most abundant in the T. aoutii holobiont (Figure 2). Together, they accounted for 80.9% of CEs in the host (174 modules) and 46.5% in the microbiome (47 modules). AAs - redox enzymes that often act synergistically with GHs, were represented by seven families (Figure 2), accounting for 33 modules in the host and 54 modules in the microbiome. PLs in the T. aoutii holobiont belonged to eight different families and were the least abundant class, with 29 modules present only in the microbiome.
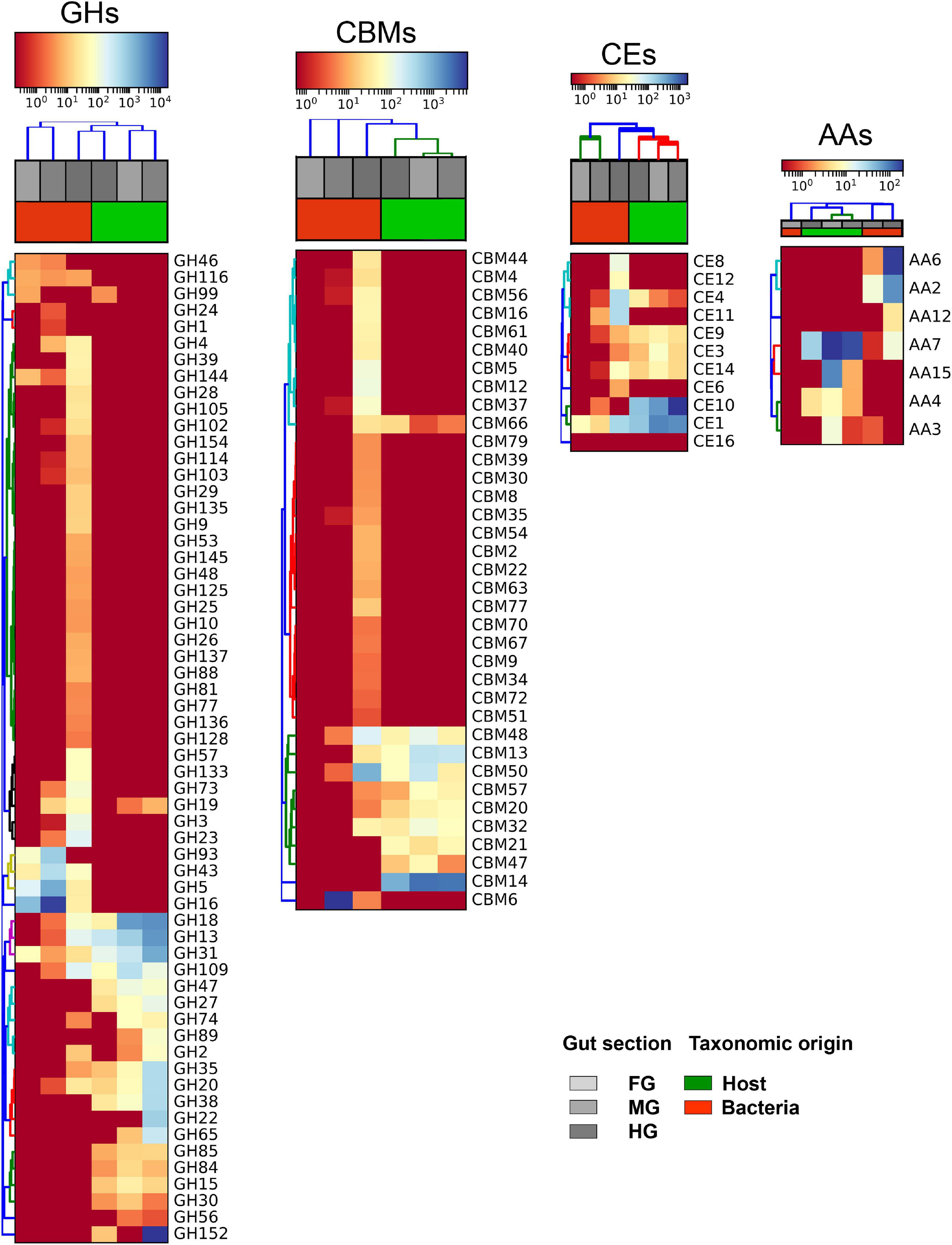
Figure 2. Heat maps of CAZy families, expressed in the host and prevailing microbiota group (Bacteria), and their distribution among the gut segments in the holobiont. The four most abundant CAZyme classes including digestive enzymes (i.e., GHs, CBMs, CEs, and AAs) were selected. TPM values were used to analyze the expression level of individual CAZyme families. Dark red and dark blue denote the lowest and highest expression, respectively. Expressed TPM value was log-transformed. Hierarchical clustering was done using Bray-Curtis dissimilarity matrices.
The most abundant digestive CAZymes (i.e., the AA, CBM, CE, and GH families) had a tissue-specific conserved taxonomic origin (Figure 2). In hierarchical clustering, CAZymes originating from the host and bacteria were clustered separately. In general, the expression of these CAZymes by bacteria was higher in HG, while FG and MG were the primary sites for the endogenous CAZymes. However, some of the CAZymes such as CBM6, GH16, GH43, GH46, GH5, and GH93 were expressed in higher numbers by bacteria in FG and MG than in HG. Compartmentalization of digestive processes plays an important role in arthropod digestion (Terra and Ferreira, 1994). Different intestinal sections play different roles in digestive processes. In T. aoutii holobiont, the differences in the distribution of identified CAZy families among holobiont parts reflected the overall diversity of expressed genes. The highest number of unique CAZy families (57 in total) was observed in the hindgut reflecting the wide diversity of genes expressed by the hindgut bacterial community (Figure 3A). More than 50% of CAZy families were found only in the microbiota, and of these, 55% of families were found only in the hindgut (Figure 3B). The fact that only 6% of the families were found together in all gut sections indicates that sections appear to differ in the transcription of CAZy microbial families. In contrast, the host data reflected one arthropod species, and a comparison of the composition of CAZy families in the different body parts showed a high degree of uniformity. More than 70% of the detected CAZy families were found in all body parts of the host (Figure 3C). The expression of many CAZymes in the non-intestinal tissues is probably related to the non-digestive functions of some enzymes. Even amylolytic enzymes may be active in the hemolymph or other body parts and play a non-digestive role in insects (Da Lage, 2018). In terrestrial isopods, the role of some CAZymes in cuticle sclerotization has been discussed (Bredon et al., 2018). The expression of potentially ligninolytic CAZy families in the non-digestive tissues of T. aoutii can be explained by the presence of activities such as ecdysone oxidase (EC 1.1.3.16), known to belong to the AA3_2 subfamily, and (R)-mandelonitrile oxidase (EC 1.1.3.49), which is annotated as a member of the same subfamily in the holobiont of T. aoutii. The latter, characterized in the polydesmid millipede Chamberlinius hualienensis, is involved in the production of the defense chemical benzoyl cyanide (Ishida et al., 2016). The superoxide dismutase (SOD) and tyrosinase (hemocyanin), both expressed in the T. aoutii holobiont, play a non-digestive role in tissues (Melov et al., 2000; Olianas et al., 2005).
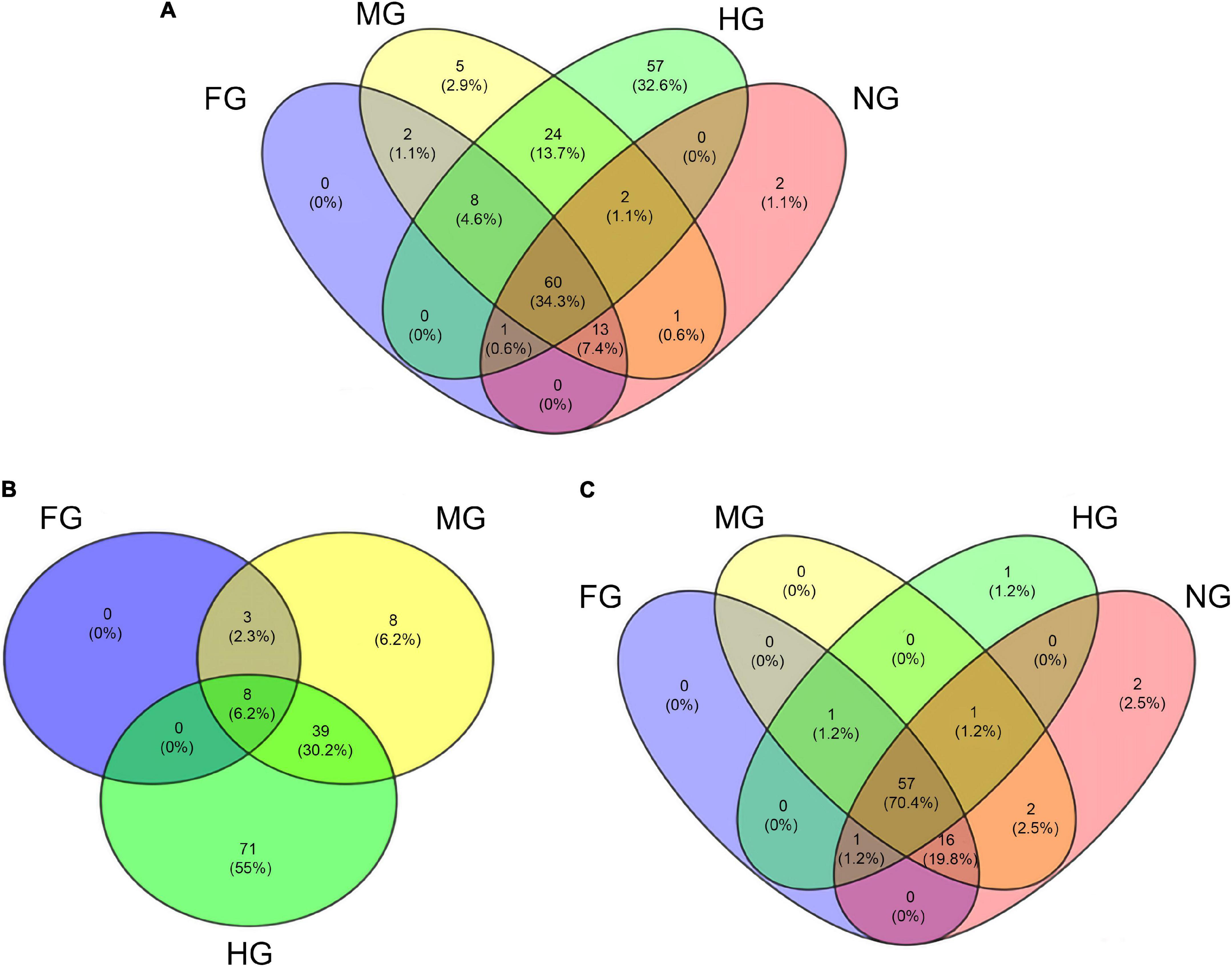
Figure 3. Venn diagram of the distribution of all CAZy families in all body parts of the T. aoutii holobiont (A), the distribution of CAZy families found in microbiota between different intestinal sections (B), and the distribution of CAZy families detected in the host between different body parts (C).
The foregut is a short segment of the intestine in millipedes (Hopkin and Read, 1992). Transport of food through the foregut is rapid, resulting in low diversity of microbial CAZymes here. It is known that the salivary glands, analyzed as part of the foregut, are the site of possible production of endogenous digestive enzymes (Fontanetti et al., 2015). However, this was not reflected in our results, as the diversity of expressed endogenous CAZymes in the foregut was relatively low. The midgut of T. aoutii had the greatest diversity of endogenous CAZymes, consistent with previous findings on the digestive physiology of millipedes (Hopkin and Read, 1992; Šustr et al., 2020a). The hindgut is the hotspot for microbial CAZyme production in the holobiont of T. aoutii.
In addition to the host, gut bacteria were the major producers of CAZymes in the holobiont (Supplementary Figure S2). Proteobacteria and Bacteroidetes were the major CAZyme producers in the millipede gut, followed by Firmicutes. Considering the extent of involvement of GHs in carbohydrate degradation, we specifically examined the bacterial phyla involved in the expression of different GH families in the hindgut and midgut (Figure 4). The association map illustrated the increased diversity of the active bacterial community and the increase in the number of CAZymes produced in the hindgut. Proteobacteria dominated and produced the majority of the GH families in the midgut (most produced families GH99, GH73, GH24, and GH4) and Bacteroidetes in the hindgut (most produced GH43 and GH109). Firmicutes showed higher CAZyme expression in the hindgut and produced exclusively the potentially cellulolytic families GH9 and GH48. GH13 was produced by Bacteroidetes, Firmicutes, and Proteobacteria in the hindgut and only by Proteobacteria in the midgut. In the hindgut, Lentisphaerae, Tenericutes, and Synergistetes produced GH families in addition to the taxa mentioned above. Bacteria play a very important role in the production of CAZymes in the millipede gut, especially in the hindgut. This agrees with a rich bacterial community previously reported in the intestinal tract of millipedes (Byzov, 2006). Within prokaryotes, Bacteria are potentially more important producers of GHs than the Archaea (Geng et al., 2021). In the millipede intestine, Archaea are mainly involved in methanogenesis as a final step of the fermentation processes (Horváthová et al., 2021).
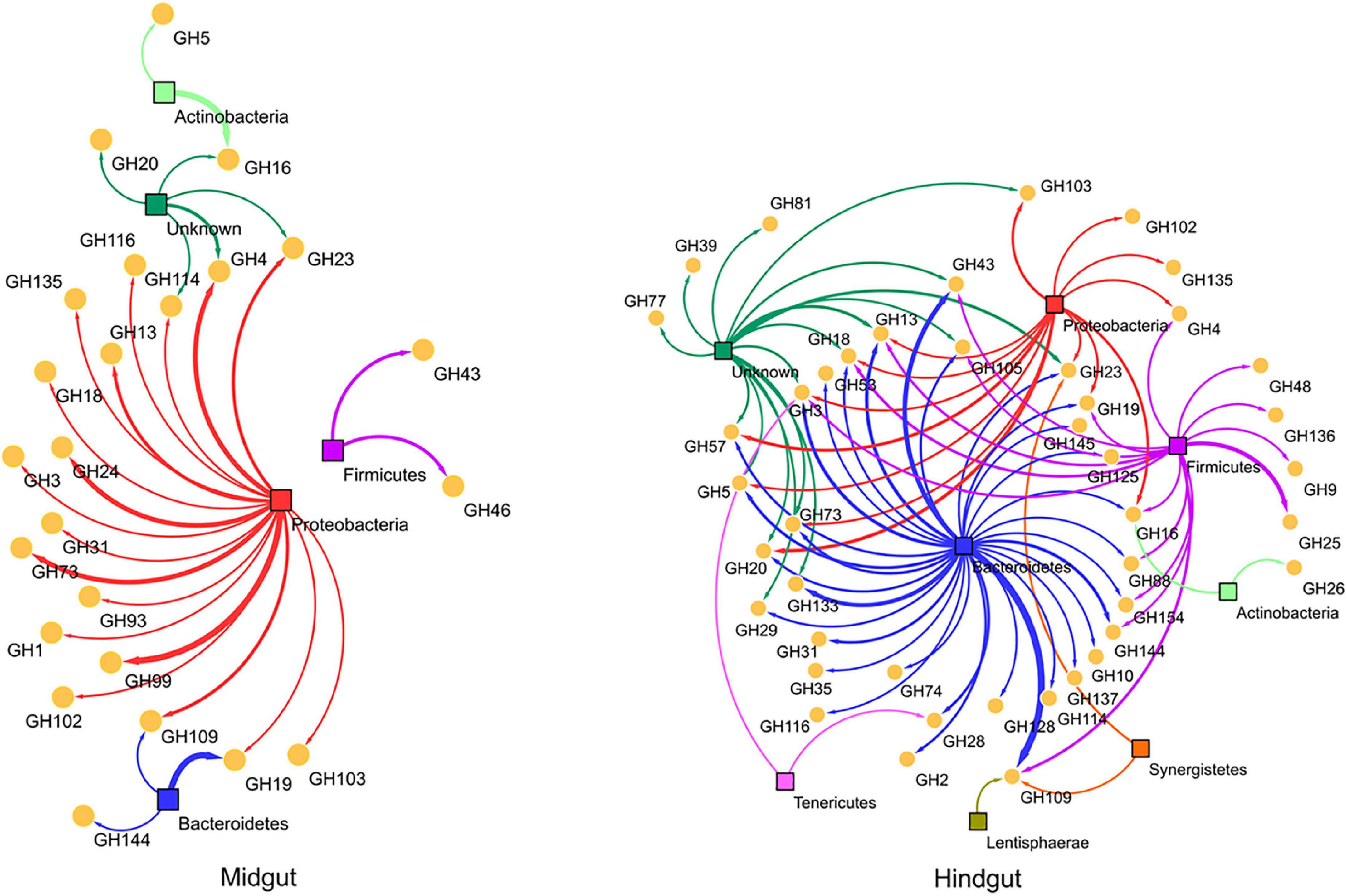
Figure 4. An association map to describe the qualitative and quantitative differences in expressed GH families between different bacterial phyla. Bacterial phyla and GH families were shown in colored squares and yellow spheres, respectively. The thickness of the connecting lines is proportional to the number of representative contigs of GH families in the phyla.
The CAZy families containing lignocellulolytic and digestive enzymes and their distribution among all parts of the holobiont and major taxa are listed in detail in Supplementary Table S2. In addition to dbCAN, a total of 39,360 transcripts with recognized EC numbers in the holobiont were classified using eggNOG. EC numbers involved in lignocellulolytic degradation and digestion are summarized in Table 1 and Supplementary Table S3. The assignment of the EC numbers discovered in the metatranscriptome to the CAZy families expressed in the holobiont is shown in Supplementary Table S4.
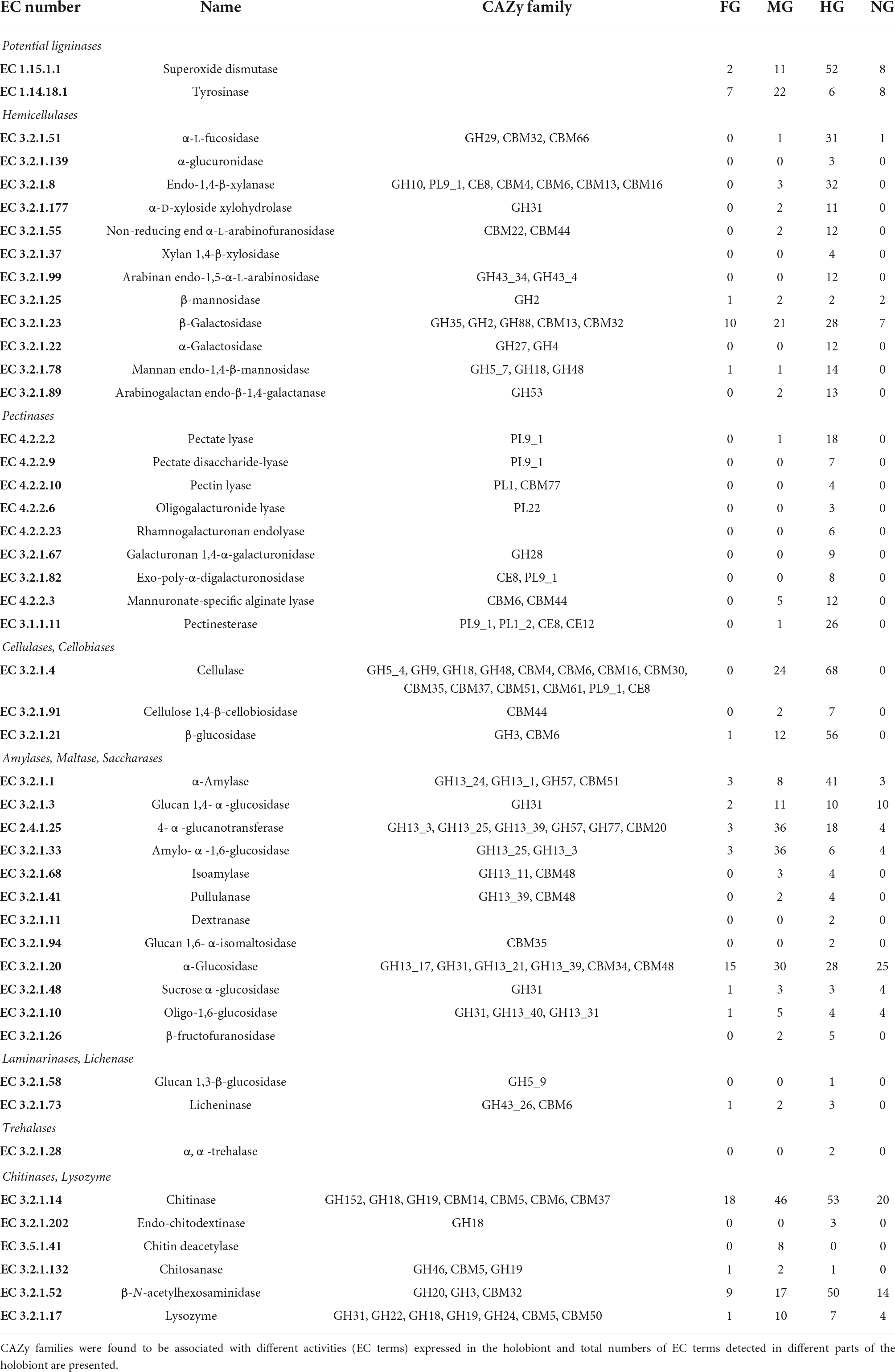
Table 1. List of CAZymes involved in the degradation of dietary important saccharides in the T. aoutii holobiont.
The presence of enzymes with different substrate specificities in the same CAZy family partially complicates the use of CAZy families as functional indicators of enzymatic activity, but an applicable functional classification of CAZy families is possible, as shown by Geng et al. (2021), who classified GHs based on primary enzyme functions. In this study, CAZy families containing EC activities corresponding to lignocellulolytic and digestive enzymes, expressed in the T. aoutii holobiont, were classified according to the presence of enzyme activities involved in the degradation of different groups of saccharide substrates. This specific functional classification resulted in 20 groups of CAZy families including enzymes that cleave starch, glycogen and saccharose (A), trehalose (T), hemicelluloses (H), cellulose (C), laminarin or lichenin (L), lignin (G), and pectin (P) (Supplementary Figure S3). The substrate-based classification of each of the CAZy families has been listed in Supplementary Table S2. The classification indicates a wide range of substrates that can potentially be targeted by the enzymes expressed in the holobiont. The broad spectrum of saccharolytic enzymes in the millipede intestine can be surmised based on previous measurements of enzyme activity in different millipede species (Šustr et al., 2020a). The classification allowed the following comparisons and analyses, which focused directly on nutritionally and ecologically important substrate groups.
Degradation of the lignocellulolytic complex in the millipede intestine
Intestinal CAZymes, of endogenous or microbial origin, were involved in the degradation of various components of the millipede diet. In this diet, relatively recalcitrant lignocellulose components predominated over easily digestible saccharides of plant and microbial origin. In the lignocellulolytic complex, components such as lignin, hemicellulose, and pectin protect a principal component, cellulose, from enzyme attack. Complete digestion and utilization of lignocellulose require at least partial disruption of these components. The ligninolytic, hemicellulolytic, and pectinolytic potential of intestinal enzymes is discussed for the first time.
The expression of the enzymatic machinery for lignin degradation was not confirmed
The lignin modifying enzymes (LMEs) are rare in animals (Bredon et al., 2018). However, the presence of LMEs has been previously demonstrated in termites (Ni and Tokuda, 2013) and terrestrial isopods (Bredon et al., 2018), whereas they are absent in the millipede T. aoutii. Only two CAZy subfamilies with potentially ligninolytic EC activities were found in the metatranscriptome data of T. aoutii holobiont. AA3_2 was expressed in the host, mainly in non-intestinal tissues. The same family was detected in midgut bacteria. AA2 was expressed in the midgut and mainly in the hindgut bacterial community (Figure 5A). Bacterial ligninases were expressed in the hindgut of T. aoutii as members of the AA2 family. However, the actual activity of LMEs in the hindgut of millipede is questioned by the fact that low oxygen levels prevail here (Horváthová et al., 2021), as in the caeca of isopods (Bredon et al., 2018) or in the P3 lumen fluid of termites (He et al., 2013). The minor role of aerobic fungi, usually capable of lignin degradation, in the holobiont, corresponds with oxygen limitation. The low expression frequency of ligninolytic CAZy families was shown in Supplementary Figure S4. Expression of the ligninolytic enzymes known to be associated with AA2 (lignin peroxidase, manganese peroxidase, versatile peroxidase) and AA3_2 (glucose oxidase, aryl alcohol oxidase) was not confirmed in T. aoutii holobiont by detection of EC numbers. Both families can be associated with non-ligninolytic activities, such as general peroxidases EC 1.11.1.-. Most AA2 entries, were linked to EC 1.11.1.21 and EC 1.11.1.5. All AA3_2 modules were linked to EC 1.1.3.49 (in the host) or EC 1.1.91.1 (in bacteria). Two potentially ligninolytic enzymes were expressed in the holobiont of T. aoutii. SOD (EC 1.15.1.1) was expressed in all host tissues, in bacteria, particularly in the hindgut, and, to a lesser extent, in ciliates. Tyrosinase (EC 1.14.18.1) was mainly expressed in the host (Table 1). SOD was discussed by Rashid et al. (2015) as ligninolytic. However, this enzyme usually plays other physiological roles. Tyrosinase showed similarities to hemocyanin (Olianas et al., 2005), the respiratory protein of millipedes (Damsgaard et al., 2013). Annotation identified tyrosinase activity in T. aoutii with hemocyanin. The role of hemocyanin in lignin degradation has been discussed in crustaceans (Besser et al., 2018). Both enzymes were also expressed in non-intestinal tissues of T. aoutii suggesting a role beyond lignin degradation, such as a laccase gene in A. vulgare isopod (Bredon et al., 2018).
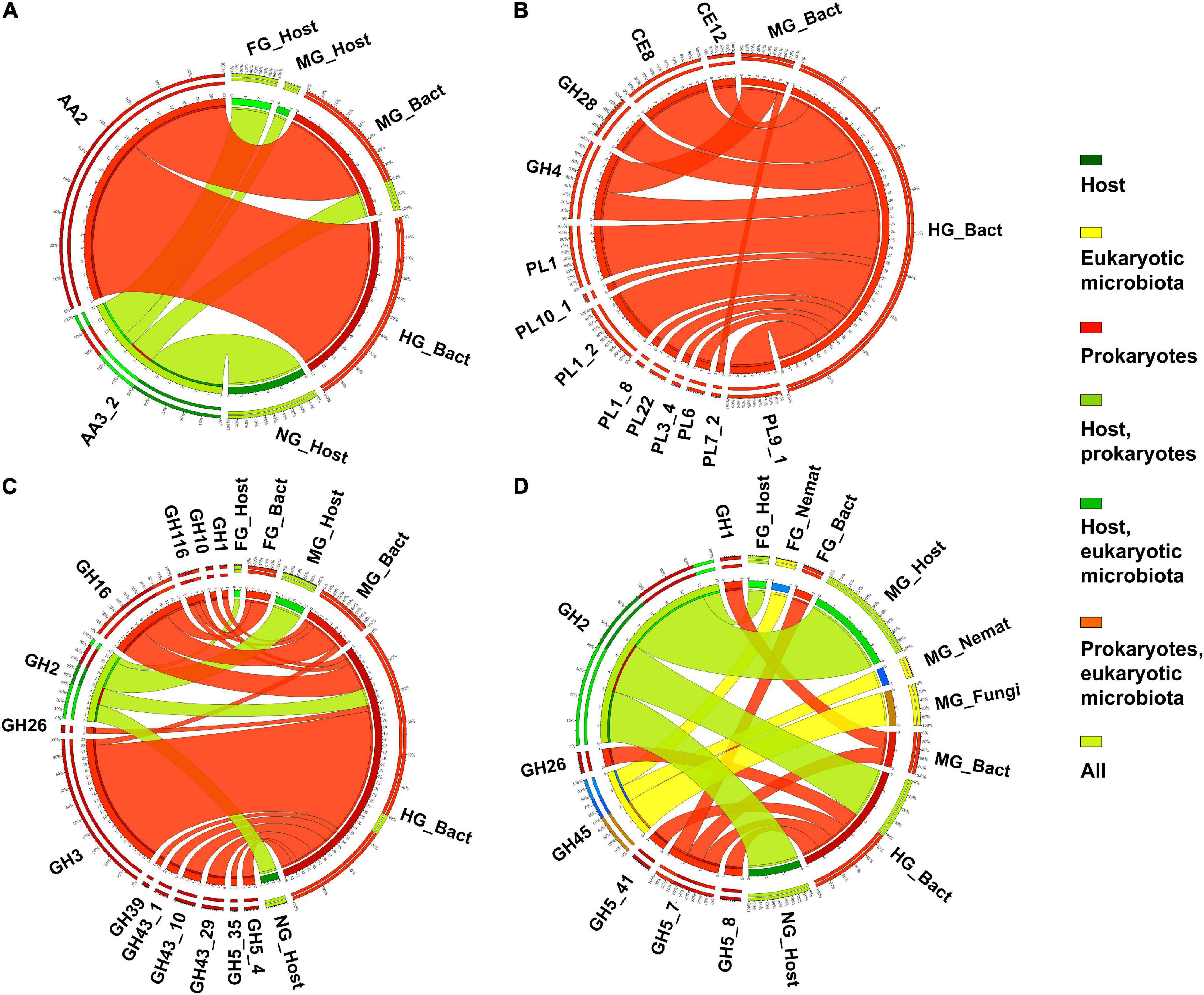
Figure 5. The distribution of expressed CAZy families containing ligninases (A), pectinases (B), xylanases (C), and mannanases (D) (as verified in the CAZy database) in the microbiota and host in different body parts. Nemat, nematoda; Bact, bacteria; Archa, archea; and Cilio, ciliophora.
Pectinases were mainly expressed in the hindgut bacteria
Pectin is regularly deposited in the secondary growth of plants. It represents a store of galactose sugars and essential minerals (e.g., calcium) (Kohn and Luknár, 1977). Pectinolytic enzymes are rarely found in animals. Pectin degradation in millipede homogenates has been confirmed in only three of eight millipede species studied (Nielsen, 1962; Marcuzzi and Turchetto Lafisca, 1976; Nunez and Crawford, 1976; Beck and Friebe, 1981). Fungal transcripts, predicted to encode GH28 (polygalacturonases), were expressed in the beetle A. glabripennis (Scully et al., 2014). The fungal and bacterial pectinase genes were found in the genome of some insects, suggesting horizontal gene transfer (Shelomi et al., 2016).
Twelve different potentially “pectinolytic” CAZy families were expressed in intestinal bacteria, most of them in the hindgut. In the midgut, only the GH4 and PL7_2 families were expressed (Figure 5B). The host does not express autochthonous CAZy families or EC terms. The microbial pectinases were mainly expressed in the hindgut (Supplementary Figure S5). The hindgut bacteria expressed proteins with the following activities of: (i) pectinesterase (EC 3.1.1.11), identified as members of CE8 and CE12 families and PL9_1 and PL1_2 subfamilies; (ii) pectate lyases (EC 4.2.2.2) belonging to PL1 and PL9 and associated with CBM37 and CBM77; (iii) pectate disaccharide-lyases (EC 4.2.2.9, PL9); (iv) pectin lyases (EC 4.2.2.10, PL1 and CBM77); (v) galacturonan 1,4-α-digalacturonosidase (EC 3.2.1.67, GH28); and (vi) oligogalacturonide lyases (EC 4.2.2.6, PL22) (Table 1 and Supplementary Tables S3, S4).
Expression of a broad spectrum of hemicellulolytic enzymes in the holobiont
The composition of hemicellulose is variable and differs between different taxonomic groups of plants, and between specific plant tissues (such as primary and secondary cell walls) (Albersheim et al., 2011). The complicated chemical structure of hemicellulose is reflected in the wide diversity of hemicellulolytic enzymes that cleave it. Consistent with this diversity, the variability of CAZy families containing hemicellulases expressed in T. aoutii was enormous. These families largely overlap with families containing other activity types. They are generally expressed in hindgut bacteria but have also been detected in the host midgut and in the eukaryotic microbiota in the midgut and hindgut (Figures 5C,D). The highest expression diversity of hemicellulolytic activity was found in the bacterial hindgut community of the T. aoutii holobiont (Supplementary Figure S6). The backbone of hemicellulose (xyloglucans and galacto-glucomannans) can be cleaved by endo-1, 4-β-xylanases (EC 3.2.1.8), and mannan endo-1,4-β-mannosidases (EC 3.2.1.78), which were expressed almost exclusively by the bacterial hindgut community (Figure 6). Some of the xylanases were directly identified as members of GH10 (Table 1 and Supplementary Table S4) and were associated with CBMs 4, 6, 13, and 16. The mannan endo-1,4-β-mannosidases are partially identified as GH5. α-D-xyloside hydrolases (EC 3.2.1.177), expressed in hindgut bacteria of T. aoutii, were identified as members of the GH31 family. The xylanolytic activity was previously tested in homogenates of the digestive tract of nine millipede species (Nielsen, 1962; Nunez and Crawford, 1976; Beck and Friebe, 1981; Urbášek and Tajovský, 1991; Cazemier et al., 1997; Šustr et al., 2020a). The presence of xylanase was confirmed in six of nine species tested, including two members of the Spirostreptidae, Orthoporus ornatus, and one undetermined species (Nunez and Crawford, 1976; Cazemier et al., 1997). However, xylan degradation was not measurable in the spirostreptid A. gigas (Šustr et al., 2020a). Hydrolysis of xylan was carried out in the isopod A. vulgare by xylanases (belonging to family GH10) and xylosidases (belonging to families GH3 or GH43) of bacterial origin (Bredon et al., 2018). Xylanases showed weak activity against xyloglucans, but xyloglucan-specific endo-β-1,4-glucanases (EC 3.2.1.151) mainly hydrolyze the xyloglucan backbone (Tokuda, 2019). However, this activity was not directly confirmed in the terrestrial isopod (Bredon et al., 2018), or in T. aoutii (this study). On the other hand, endo-mannanases (EC 3.2.1.78) are expressed in the microbiome of T. aoutii, but not in A. vulgare (Bredon et al., 2018).
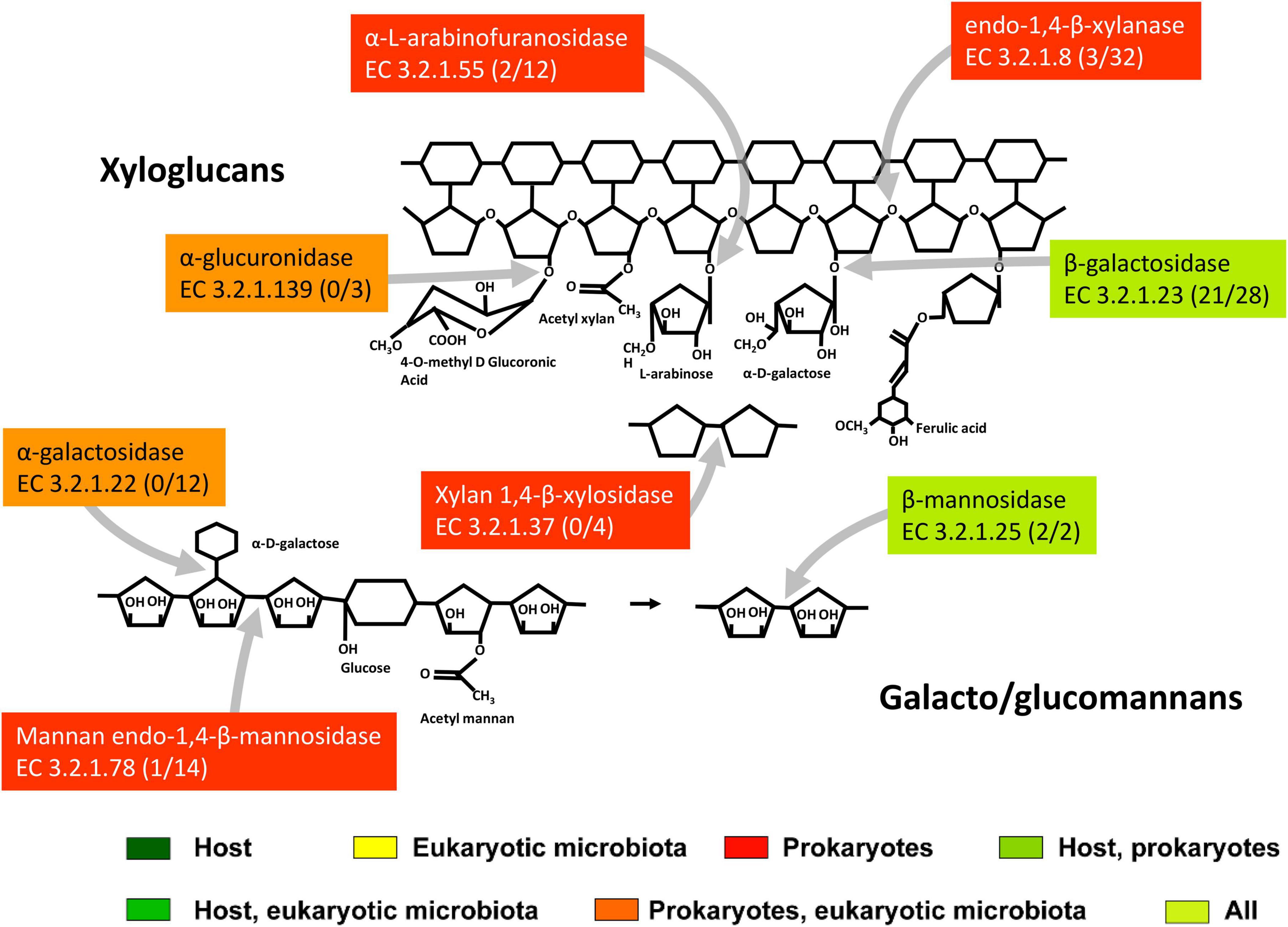
Figure 6. The scheme of action of main hemicellulases expressed in the midgut and hindgut of the millipede T. aoutii. Frequencies of enzymes in midgut/hindgut are presented in parenthesis following the EC number. Gray arrows – types of bonds attacked by different types of enzymes. The basic scheme was modified from Ulaganathan et al. (2015).
The disaccharides xylobiose and mannobiose formed by cleavage of the hemicellulose backbone can be further cleaved in T. aoutii by xylan 1,4-β-xylosidases (EC 3.2.1.37) expressed by bacteria and by endogenous or bacterial β-mannosidases (EC 3.2.1.25). Some of the endogenous mannosidases are classified as GH2. The isopod A. vulgare has endogenous β-mannosidase (Bredon et al., 2018) of the GH2 family, as does T. aoutii.
The hemicellulose backbone has side chains, containing mainly arabinose and glucuronic acid residues (Ordaz-Ortiz and Saulnier, 2005). Enzymes that attack the side chains of hemicellulose, β-galactosidases (EC 3.2.1.23) of GH35 and GH2, α-L-fucosidases (EC 3.2.1.51) of GH29, α-galactosidases (EC 3.2.1.22, GH4), α-glucuronidases (EC 3.2.1.139) and α-L-arabinofuranosidase (EC 3.2.1.55) were expressed in bacteria in the holobiont of T. aoutii. Arabinans can be targeted by bacterial arabinan endo-1,5-α-L-arabinosidase (EC 3.2.1.99, GH43). The host expresses β-galactosidases (GH35) and α-L-fucosidases. The expression of α-galactosidases and α-glucuronidases was found in the eukaryotic microbiota of T. aoutii. β-Galactosidases were frequently expressed in holobionts of soil detritivores (in A. vulgare endogenous GH33 and in the microbiome GH2 and GH42, Bredon et al., 2018). In A. vulgare, α-L-arabinofuranosidases (EC 3.2.1.55, of microbial origin from GH51 or GH43) were expressed (Bredon et al., 2018). In this isopod, endogenous α-galactosidases from GH27 were complemented by microbial enzymes from GH4 and GH36. In addition, the microbiota of terrestrial isopods produced acetyl-xylan and feruoyl esterases, which extend the ability to cleave hemicellulose side bonds (Bredon et al., 2018).
Cellulases were mainly expressed in the hindgut bacteria
Previous measurements confirmed the degradation of some β-glucosides, carboxymethyl, or crystalline cellulase by intestinal homogenates of some millipede species. Low β-glucosidase and cellulase activities were detected in spirobolid E. pulchripes and spirostreptid A. gigas, with acidic pH optima corresponding to the midgut (Šustr et al., 2020a). In these two species, cellulases were detected not only in the hindgut but also in the midgut (including the intestinal wall).
However, our metatranscriptomic data showed higher expression of cellulolytic genes in the hindgut of T. aoutii. CAZy families associated with potentially cellulolytic activities were diverse in the holobiont and reassociated with many other types of activities. The distribution of all families containing exo- or endo-glucanases is shown in Figure 7A. These families were mainly expressed in intestinal prokaryotes (GH3 dominated). GH45 was expressed by the eukaryotic microbiota of the foregut and midgut, and GH74 was expressed not only by bacteria but also by the host. The GH74 family may include endoglucanases and xyloglucanases. The expression of this family was previously described in the isopod A. vulgare Bredon et al. (2018). However, in T. aoutii, expression of this family is associated with cellulolytic function only in bacteria in the hindgut. The next potentially cellulolytic family, AA15, was endogenous. Of the families known to be associated with cellobiases (Figure 7B), only GH30_1 was endogenous. Intestinal bacteria expressed GH2 in addition to their host. Other prokaryotic genes represented six families dominated by GH3 and GH16. The fungal community expressed subfamily GH5_9 in the hindgut. Cellulolytic bacterial CAZy families and EC numbers were expressed mainly in the hindgut and to a lesser extent in the midgut. Analysis of the data at the EC-number level did not confirm the expression of endogenous cellulases (Supplementary Figure S7).
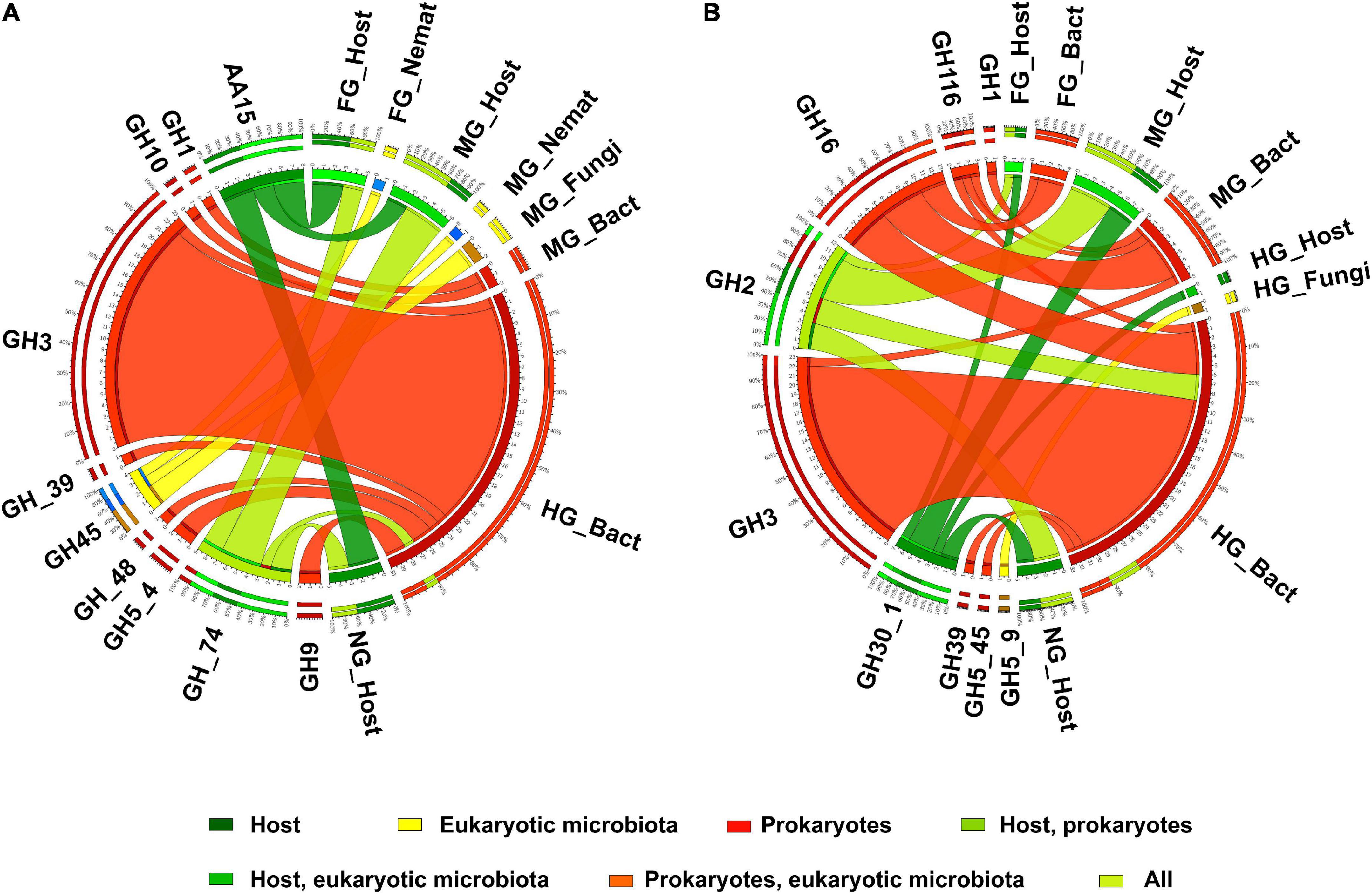
Figure 7. The distribution of expressed CAZy families including endo- or exo-glucanases (A), cellobiases (B), (as verified in the CAZy database) in the microbiota and in the host in different body parts. Nemat, nematoda; Bact, bacteria; Archa, archea; Cilio, ciliophora.
Endogenous lytic cellulose monooxygenase (EC 1.14.99.54), known to belong to the AA15 family and expressed in the host of T. aoutii, could not be confirmed at the level of EC numbers in the holobiont. In the midgut of the firebrat Thermobia domestica, a lytic cellulose monooxygenase from the AA15 family oxidizes C1 sites to cleave β-1,4-glycosidic bonds, which increases the activity of cellulolytic and chitinolytic enzymes (Sabbadin et al., 2018). LMPOs belonging to AA15 are widely distributed among insects (Sabbadin et al., 2018; Ceja-Navarro et al., 2019), but their role in lignocellulose degradation remains unclear and further studies are needed (Tokuda, 2019). Other endogenous cellulases, notably GH5, GH9, and GH45, have been detected in several Mandibulata species, and the ancestors of Protostomia are thought to have possessed these genes (Watanabe and Tokuda, 2010). The above GH families, known to be associated with endogenous animal cellulases (Watanabe and Tokuda, 2001, 2010; Cragg et al., 2015), were expressed by the gut microbiota only in the holobiont of T. aoutii. The lack of expression of endogenous cellulases or β-glucosidases in T. aoutii is in contrast to results from terrestrial isopods (Bredon et al., 2018).
The first step of cellulose degradation in the hindgut of T. aoutii may be covered by expressed endoglucanases (cellulases, EC 3.2.1.4) and exoglucanases (cellulose 1,4-β-cellobiosidases, EC 3.2.1.91), both of prokaryotic origin (Figure 8 and Supplementary Figures S8–S10). Transcripts corresponding to these activities were more abundant in the hindgut than in the midgut. Bacterial endoglucanases were associated with GH5_4 and GH9. Enzymes with functionality EC 3.2.1.4 expressed by eukaryotic microbiota, can probably be associated with GH26, GH74, or GH45. The GH45 family expressed in nematodes and fungi in the holobiont of T. aoutii has been horizontally transferred from fungi to plant-parasitic nematodes (Palomares-Rius et al., 2014). Proteins of this family can be expressed by fungi or nematodes that originally live in soil and are ingested with food. Bacterial genes encoding EC 3.2.1.4 associated with several carbohydrate-binding modules (CBM 4, 6, 16, 30, 35, 37, 51, and 61) were mainly active in the hindgut. Exocellulases (cellulose 1,4-β-cellobiosidase, EC 3.2.1.91) were expressed in hindgut bacteria with CAZy affiliation to CBM44. Cellulolytic genes, belonging to the GH5, GH9, and GH48 families associated with many CBMs were expressed in the holobiont of T. aoutii, mainly by the hindgut bacterial community. Similarly, the hindgut of lower termites, which has a community of cellulolytic protists, shows significantly higher hydrolytic activities toward microcrystalline cellulose than other gut regions and the salivary glands (Tokuda et al., 2005; Lo et al., 2011).
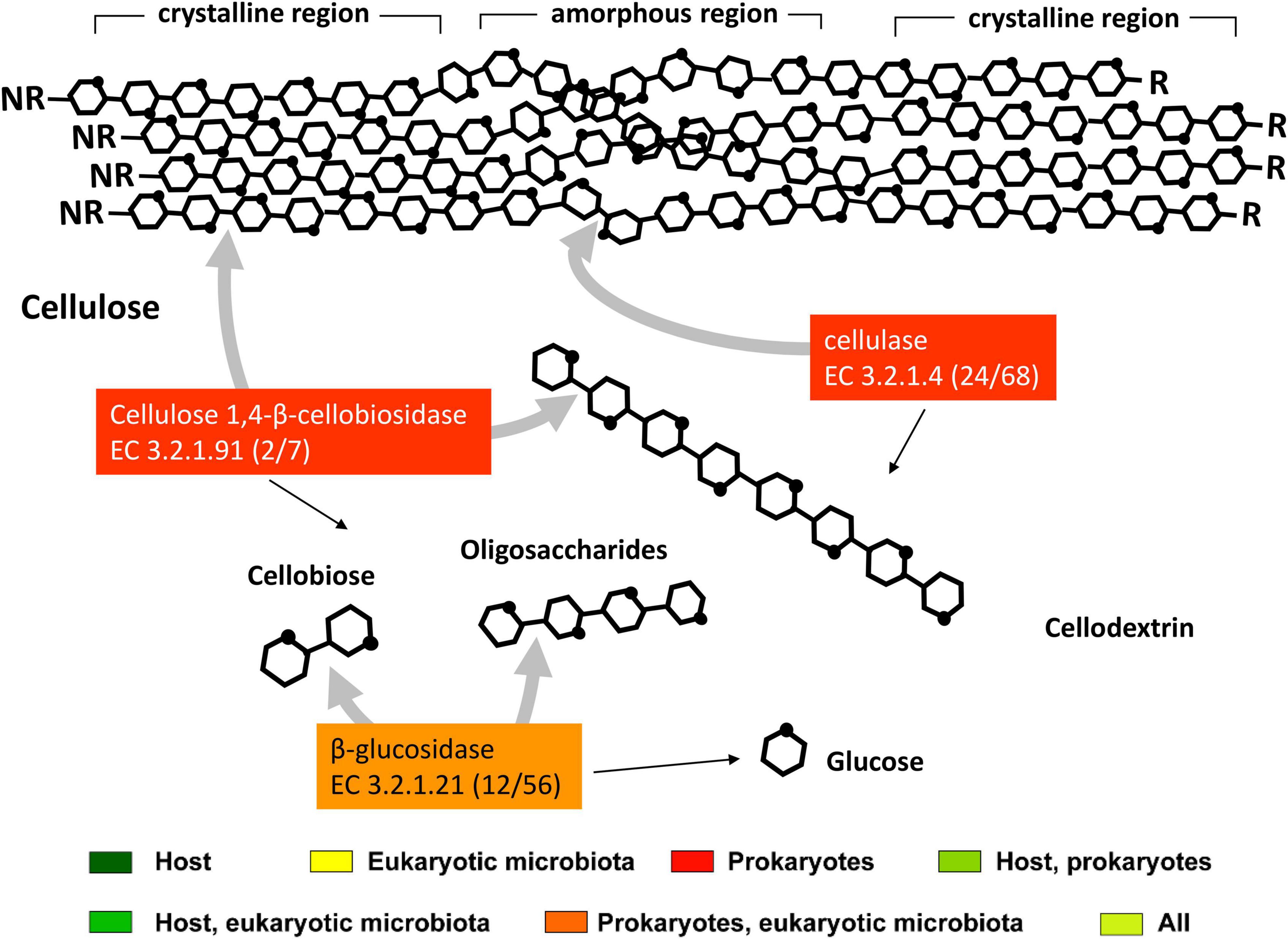
Figure 8. The scheme of action of enzymes for cellulose degradation is expressed in the midgut and hindgut of the millipede T. aoutii. A number of transcript annotations of enzymes in midgut/hindgut is presented in parenthesis following the EC number. Gray arrows – types of bonds attacked by different types of enzymes, black arrows – expected range of reaction products. The scheme of enzymatic cellulose degradation was modified by Ezeilo et al. (2017), using information from Singh et al. (2016) and KEGG db (Kanehisa and Goto, 2000).
Expression of bacterial β-glucosidases (EC 3.2.1.21) was detected in all three intestinal sections, with the highest variability in the hindgut. β-Glucosidases were associated with the GH3 and CBM6 families (Table 1 and Supplementary Tables S3, S4). In addition, enzymes with this type of activity can be assigned to families GH16, GH2, GH116, and subfamily GH5_45 expressed in bacteria. The fungal community expressed some cellobiases with unknown CAZy affiliation in the hindgut. The expression of endogenous β-glucosidases was not confirmed in this study in T. aoutii and in the isopod A. vulgare (Bredon et al., 2018). In both holobionts, the enzyme was expressed by the microbiome (as GH3 and GH1 proteins for T. aoutii and A. vulgare, respectively). In addition, a β-glucosidase of unknown family affiliation is expressed by hindgut fungi in the holobiont T. aoutii. However, in general, β-glucosidases are widely expressed in animals (Byeon et al., 2005). In termites, β-glucosidases play other functions besides digestion, including egg recognition and social communication (Matsuura et al., 2009; Shimada and Maekawa, 2014). These β-glucosidases have been associated with GH1 (Terra and Ferreira, 2005), with the exception of a GH3 from the midgut of Hodotermopsis sjostedti (Yuki et al., 2008). The holobiont T. aoutii expressed GH30_1 and GH2, which are known to contain β-glucosidases. However, all counts of these families in the host were identified as β-glucosylceramidases, β-galactosidases, or β-mannosidases. The effectiveness of cellulose degradation is often associated with the synergistic action of the three major cellulase types: endoglucanases, cellobiohydrolases, and β-glucosidases. While endoglucanases and β-glucosidases are found in many organisms, cellobiohydrolases are restricted to some bacteria, fungi, and protists (Watanabe and Tokuda, 2010; Lo et al., 2011). Cellobiohydrolase genes were not detected in the metatranscriptomes of some lower termites and terrestrial isopods (Dedeine et al., 2015; Bredon et al., 2018) and were present at low levels in the holobiont of T. aoutii. The absence of cellobiohydrolases could be compensated by the high number of endoglucanases and mechanical fragmentation of the food (Bredon et al., 2018). Several domains indicative of cellulosome activity were expressed in intestinal bacteria, mainly in the hindgut. Dockerin domains linking catalytic, cohesin, and SLH domains in the cellulosome were found only in the hindgut bacterial community. One dockerin was associated with Ruminococcaceae, families GH5, GH9, and GH48, the celB gene, and the endocellulase EC 3.2.1.4; the second with Peptoniphilaceae and the acm gene encoding lysozyme. Cohesin domains, some of which may be components of cellulosomes linking dockerins and scaffoldin, were found in midgut bacteria associated with Oxalobacteraceae (the pulQ gene), and in hindgut bacteria (associated with Ruminococcaceae). SLH domains, linking the cellulosome to the cell wall, were more abundant in intestinal bacteria. Expression of this module type was observed in the midgut (once) and hindgut (81 times) in a wide range of bacterial taxa including Ruminococcaceae. Some of these domains were associated with CAZymes such as xylanase, amylase, pullulanase, and pectate lyase.
Importance of non-lignocellulolytic nutrient sources for a millipede
In addition to lignocellulolytic substances, millipedes also consume other nutrient sources such as starch, sucrose, laminarin, and microbial cells. Studies examining the lignocellulolytic potential of invertebrates generally do not consider the amylolytic activity of the gut, although the assimilation of energy from starch and similar storage substances is a nutritional alternative to the utilization of lignocellulose for the host. A comparison of amylolytic and lignocellulolytic digestive potentials allows consideration of the importance of lignocellulose digestion to host energy acquisition. Amylase activity has been detected in eight of eleven millipede species studied (Nielsen, 1962; Marcuzzi and Turchetto Lafisca, 1976; Nunez and Crawford, 1976; Shukla and Shukla, 1986; Urbášek and Tajovský, 1991; Šustr et al., 2020a).
The host contributed most to the expression of amylolytic CAZy modules in the midgut, whereas amylolytic modules of bacterial origin predominated in the hindgut (Figure 9A). The expressed CAZy families known to contain amylases were clearly divided into endogenous (such as the GH13_24 subfamily) or bacterial (such as GH57 and several other GH13 subfamilies). The CAZy families known to contain α-glucosidases showed a similar expression pattern to the amylases, with the endogenous proteins expressed mainly in the midgut and the bacterial proteins in the hindgut. However, the expression of α-glucosidases containing families was not clearly separated taxonomically. GH31 was expressed in the host, in eukaryotic microorganisms, and in bacteria (Figure 9B). The expressed genes, characterized by the EC numbers corresponding to starch, glycogen, and sucrose-degrading enzymes, showed a similar distribution of CAZy families containing this type of activity (Supplementary Figure S11).
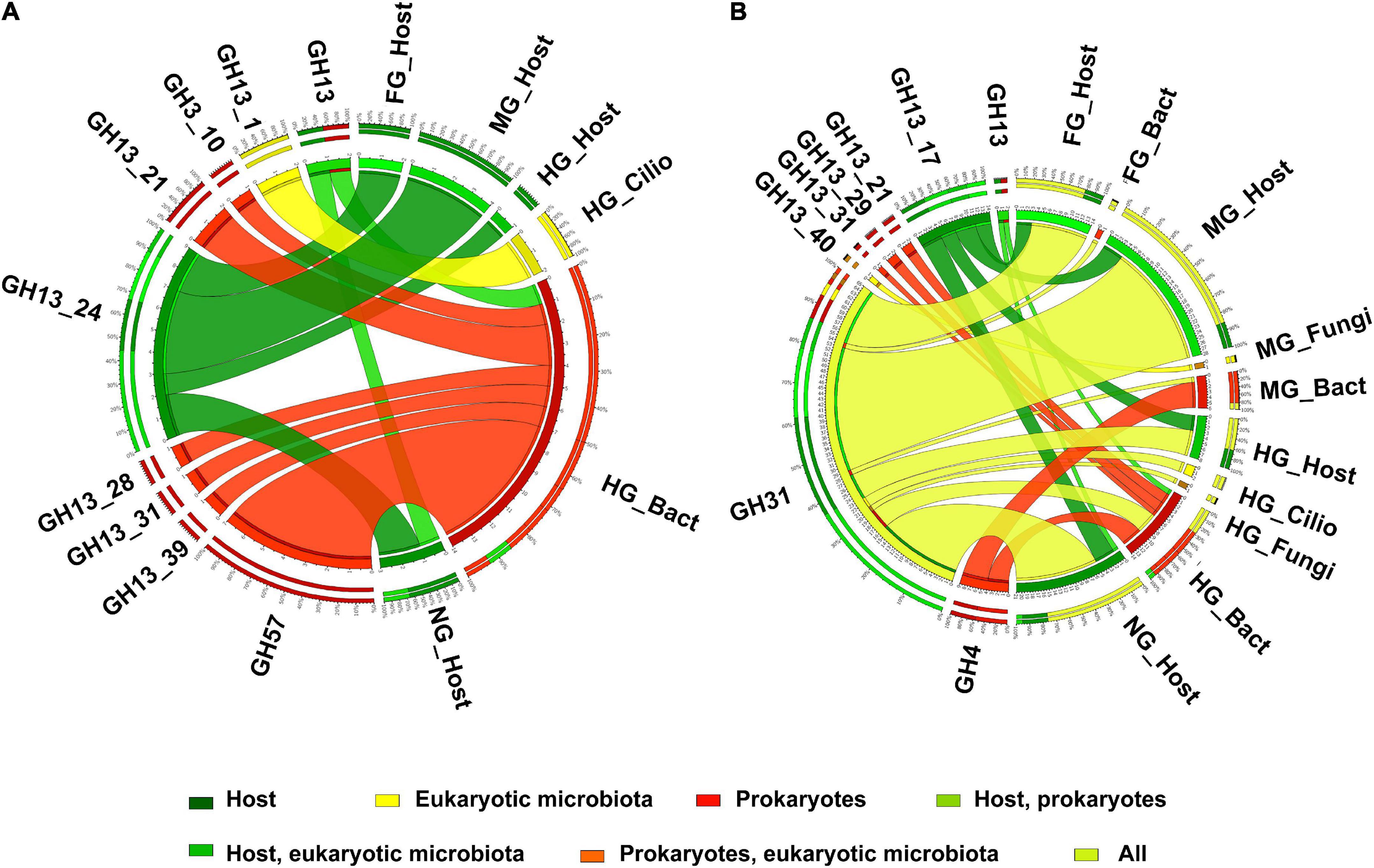
Figure 9. The distribution of expressed CAZy families including α-amylase (A) and α-glucosidase (B) (as verified in the CAZy database) in the microbiota and in the host in different body parts. Nemat, nematoda; Bact, bacteria; Archa, archea; Cilio, ciliophora.
Starch can be cleaved into maltotriose, maltose, dextrin, or glucose by endogenous α-amylases (EC 3.2.1.1) and glucan 1,4-α-glucosidases (EC 3.2.1.3) expressed in all host tissues (Figure 10, Table 1, and Supplementary Table S3). Some of the host α-amylases matched GH13, namely GH13_24 (Supplementary Table S4). α-Amylases expressed in hindgut ciliates belonged to GH13_1. GH13_1 is a subfamily containing mainly fungal amylases (Janeček and Svensson, 2022). However, a transcript from the ciliate Nyctotherus ovalis annotated in GenBank as an α-glucosidase is likely a GH13_1 α-amylase (Da Lage et al., 2007). No family has been recognized for amylases expressed in intestinal nematodes. α-Amylases are nearly ubiquitous and of paramount importance in the nutrition of bacteria, plants, fungi, and animals. Many of them possess multiple gene copies due to gene duplications or horizontal transfer (Da Lage, 2018). The GH13_24 subfamily, which includes the endogenous α-amylases in T. aoutii, is thought to contain both vertebrate amylases (Da Lage, 2018; Janeček and Svensson, 2022) and invertebrate amylases (Mills et al., 1999).
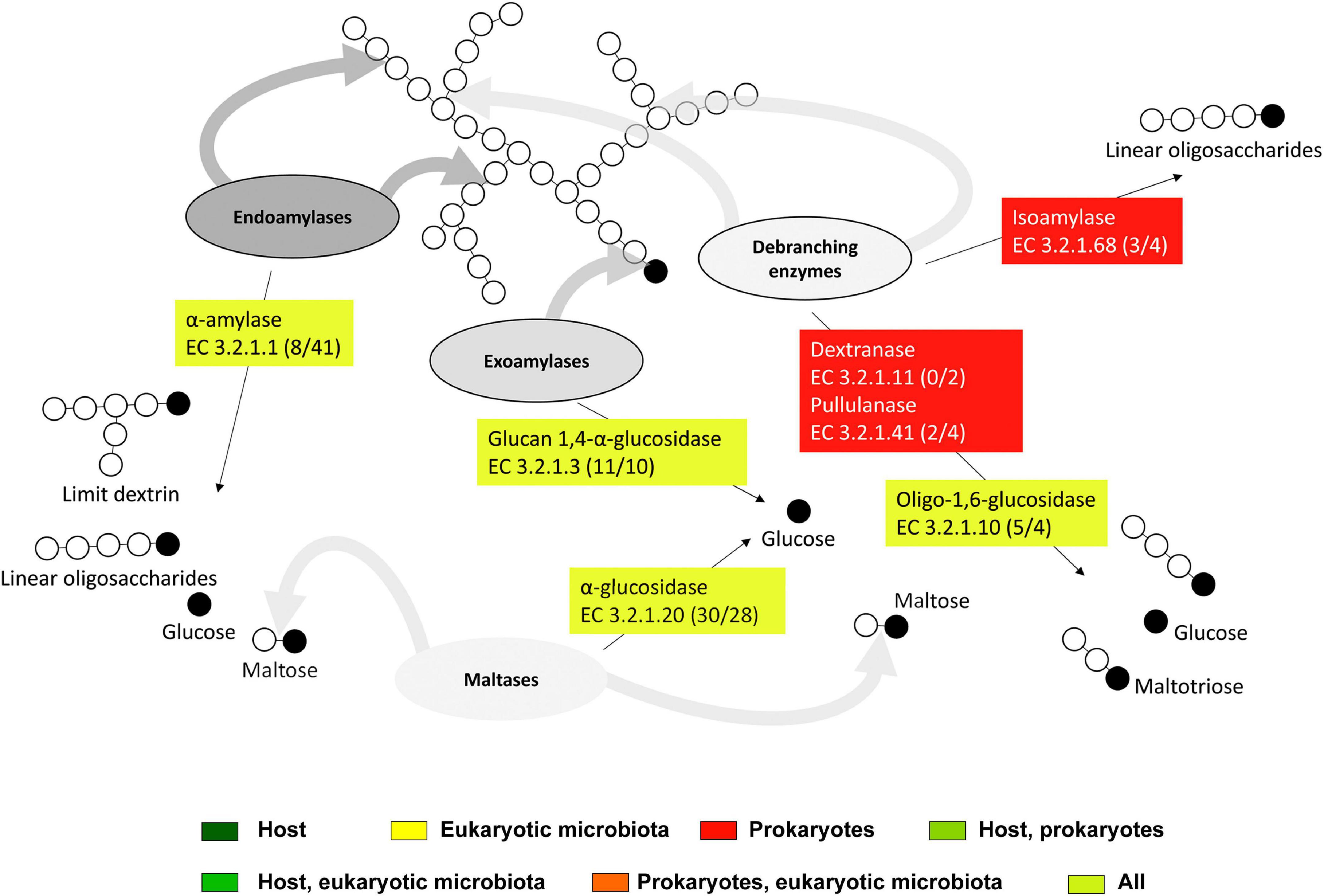
Figure 10. The scheme of action of expressed starch degradation enzymes in the midgut and hindgut of the millipede T. aoutii. Frequencies of enzymes in midgut/hindgut are presented in parenthesis after the EC numbers. Gray arrows – types of bonds attacked by different types of enzymes, black arrows – expected range of reaction products. The basic scheme was modified from Hii et al. (2012).
The bacterial α-amylases in the holobiont T. aoutii belonged to the GH57 family, and other putative bacterial amylases may belong to the GH13 family. In addition, the GH13_28 subfamily, which contains only α-amylase activity, was expressed in the hindgut bacteria of T. aoutii. Some of the bacterial α-amylases were associated with CBM51. Considering the bacterial enzymatic potential at the EC number level, the amylolytic activities expressed by the midgut bacterial community of T. aoutii are potentially sufficient for complete starch degradation, however, the diversity of CAZy amylolytic families in this section is low.
The observed family memberships of the microbial amylases are consistent with the known taxonomic ranges of the assigned families. All GH57 members are known from prokaryotes (with a ratio of ∼1:4 for Archaea:Bacteria) (Blesák and Janeček, 2012). T. aoutii expresses an endogenous γ-amylase (EC 3.2.1.3, GH31), that successively hydrolyzes terminal 1,4-α-D-glucose residues with the release of glucose from the non-reducing ends of the chains. The GH15 family, known to possess γ-amylase activity, was expressed by the host in the holobiont of T. aoutii, but all of these transcripts were annotated as GH15-like phosphorylase. The GH15 family is known in eukaryotes, particularly in fungi and some protozoa, but was also found in the transcriptome of the snout beetle (Luo et al., 2018). Most of these enzymes hydrolyze 1,6-alpha-D-glucoside bonds when the next bond in the sequence is 1,4 (Gasteiger et al., 2003). This activity confers some debranching capacity to the enzyme. Enzymes targeting side branches in amylopectin, such as isoamylase (EC 3.2.1.68) and pullulanase (EC 3.2.1.41), were expressed by microbial communities of the midgut and hindgut of T. aoutii. Bacterial dextrinase (EC 3.2.1.11) was expressed only in the hindgut. Bacterial isoamylases were identified as GH13_11, and bacterial pullulanases corresponded to GH13_39 and were associated with CBM48. Oligo-1,6-glucosidases (EC 3.2.1.10) were expressed in all host tissues, hindgut ciliates (both GH31), midgut fungi (GH13_40), and bacteria (GH13_30). The cleavage of side chains in α-polysaccharides might depend mainly on microbial enzymes.
The second step of starch digestion is represented by α-glucosidases (EC 3.2.1.20), which cleave maltose to glucose. Genes expressing α-glucosidases were detected in all body parts of the T. aoutii holobiont. High variability of endogenous α-glucosidase genes was detected in the intestine (mainly the midgut) and in non-intestinal tissues. The expressed α-glucosidases of the millipede belonged to GH31 and GH13_17. In the foregut and midgut, maltose degradation may be supported by the expression of bacterial α-glucosidases. In the hindgut, some bacterial, ciliate, and fungal α-glucosidases were expressed. The expressed bacterial α-glucosidases corresponded to GH13_21 or GH31, and the α-glucosidases expressed in ciliates and fungi were determined to be GH31 (Table 1, Supplementary Figures S8–S10, and Supplementary Tables S3, S4). GH13 α-glucosidases are mainly found in Arthropoda. There are good reasons to believe that they are ancestral to all metazoans (Gabriško, 2013). The α-glucosidases of GH13_17 and GH31 were previously reported from insects (Ohashi et al., 1996). The presence of GH31 α-glucosidases has been confirmed in bacteria, fungi, and the supergroup SAR (CAZy, see footnote 1). Some genes encoding digestive enzymes in this family are duplicated and exhibit dual specificity (Zhang and Williams, 2021). Most members of the GH31 family are multidomain proteins (da Costa-Latgé et al., 2021). In sequences from T. aoutii, GH31 activity (EC 3.2.1.20) was annotated several times along with multiple EC numbers (EC 3.2.1.20, EC 3.2.1.3, EC 3.2.1.3, EC 3.2.1.48, and EC 3.2.1.10).
It is known that α-glucosidases and oligo-1,6-glucosidases (EC 3.2.1.10) can be involved in saccharose degradation. The spectrum of saccharolytic CAZy families expressed in the T. aoutii holobiont corresponds to the distribution of families containing α-glucosidases. Regardless of the inclusion of EC 3.2.1.26 in the EC data, the expression abundances based on CAZy families and EC numbers showed similar pictures. Endogenous saccharases predominated in the midgut, whereas bacterial enzymes predominated in the hindgut (Supplementary Figure S12). The sucrase α-glucosidases (EC 3.2.1.48) of GH31 were expressed in all host tissues and hindgut ciliates. The β-fructofuranosidase type of saccharase (EC 3.2.1.26) was detected in the midgut and hindgut bacterial communities. However, their CAZy affiliation was not detected (Table 1, Supplementary Figures S8–S10, and Supplementary Tables S3, S4).
Laminarinases were detected in gut homogenates of spirostreptid and spirobolid millipedes using a chromolytic substrate (Šustr et al., 2020a). In A. gigas, laminarinolytic activity was higher in the midgut than in the hindgut. The activity in the intestinal wall of the midgut, which was crudely purified from the contents, allowed speculation about the possible endogenous origin of this enzyme (Šustr et al., 2020a). Relatively high laminarinolytic activity was detected in the midgut contents of the species studied, indicating a possible important role of β-1,3-glucan degrading enzymes in digestion. 1,3-β-D-glucans are widely distributed in plants, algae, fungi, euglenoid protozoa, and bacteria. They are involved in the cell wall structure and have a number of other biological functions (Bacic et al., 2009). Their presence in the millipede diet is likely. Laminarinases, which are thought to have a digestive function, have been found in many invertebrates (Piavaux, 1977). Families containing laminarin- and lichenin-degrading enzymes were rarely expressed by the host of the T. aoutii holobiont (family GH152 in the midgut, hindgut, and non-gut tissues, Figure 11A). Most families were of bacterial origin, especially GH3 and GH16. These families were mainly expressed in the bacterial community of the hindgut (Supplementary Figure S13). Fungal families were represented by GH5_9 expressed in the hindgut. No EC number corresponding to laminarinolytic activity was detected in the host. The putative expression of an endogenous glucan endo-1,3-beta-D-glucosidase in the millipede gut may be supported by the expression of the GH152 and GH81 families. GH152 is a long overdue family created after Sakamoto et al. (2006) showed that the enzyme from Lentinula edodes (Fungi) has β-1,3-glucanase activity. In our data, the GH152 family was detected in the host. GH81 is a family that includes only EC 3.2.1.39 activity and, according to information from the CAZy database, is common in bacteria, fungi, plants, and oomycota. At the EC-number level, expression of licheninases (EC 3.2.1.73) in the holobiont of T. aoutii was confirmed in gut bacteria (belonging to GH43). The exo-1,3-β-glucanases (EC 3.2.1.58) belonging to GH5_9 were expressed in hindgut fungi (Table 1, Supplementary Figures S8–S10, and Supplementary Tables S3, S4). The CAZy affiliation of the bacterial licheninases in T. aoutii was undetermined. The enzymes act on lichenin and cereal β-D-glucans, but not on β-D-glucans, containing only 1,3- or 1,4-bonds. Glucan-1,3-β-glucosidase acts on oligosaccharides, but very slowly on laminarinobiose (Gasteiger et al., 2003). The fungal enzyme was identified in the hindgut of T. aoutii as a member of the GH5_9 subfamily, which is known to be exclusive to fungi (CAZy, see footnote 1).
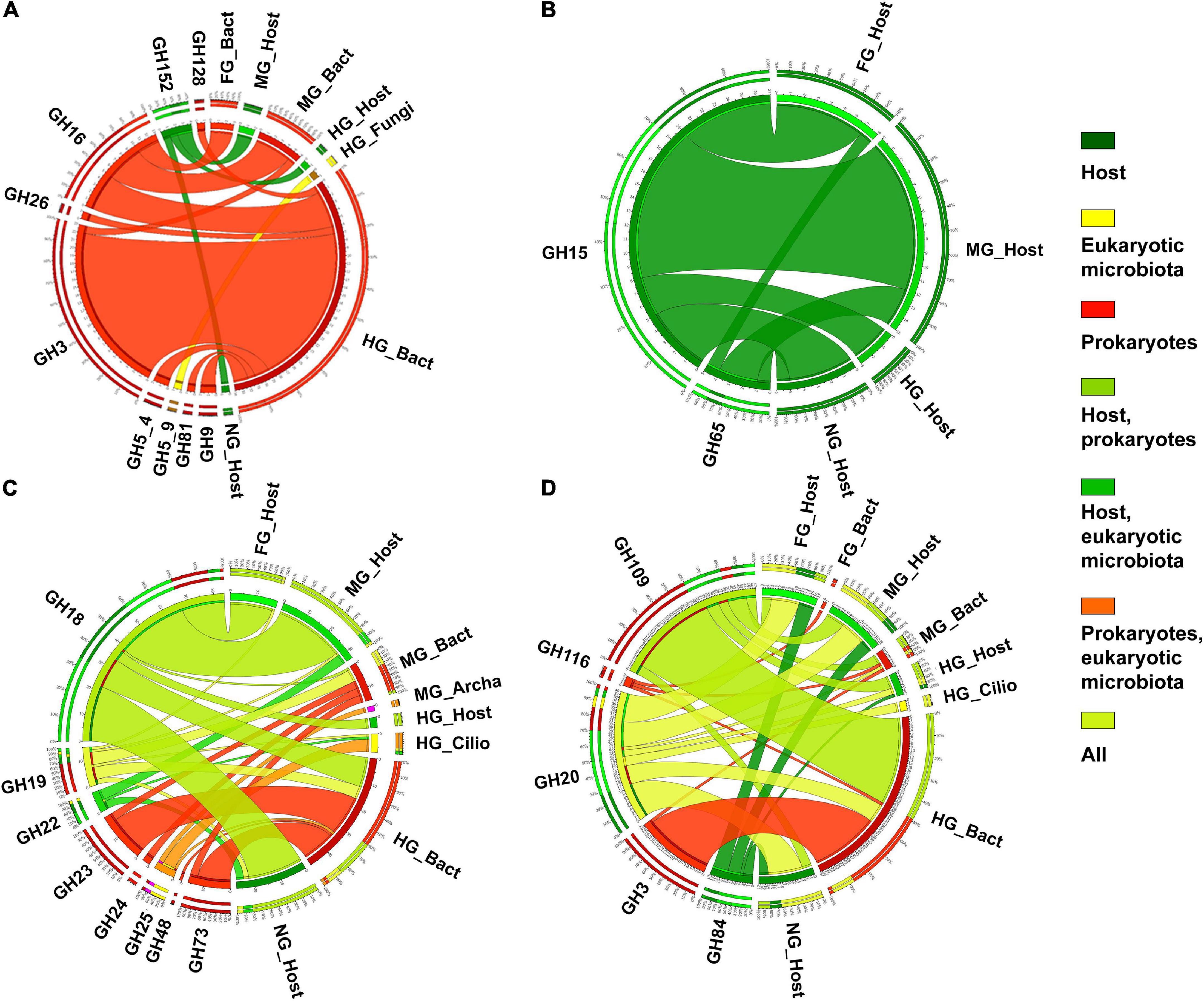
Figure 11. The distribution of expressed CAZy families containing laminarinases or licheninases (A) and trehalases (B), chitinase (EC 3.2.1.14) or lysozyme (EC 3.2.1.17) (C) and glucosaminidase (EC 3.2.1.52) (D) (as verified in CAZy database) in the microbiota and host in different body parts of the holobiont of T. aoutii. Nemat, nematoda; Bact, bacteria; Archa, archea; and Cilio, ciliophora.
The presence of trehalolytic and chitinolytic enzymes or lysozyme has been used as an indicator of trehalose and chitin digestion in fungivorous and microbivorous arthropods (Siepel and de Ruiter-Dijkman, 1993; Berg et al., 2004; Erban and Hubert, 2008). Trehalose accumulates in fungal cells (Jorge et al., 1997), fungi have high chitin content in the cell wall (Siepel and de Ruiter-Dijkman, 1993), and lysozyme hydrolyzes bacterial cell walls (Erban and Hubert, 2008). In addition, some chitinases showed some lysozyme side activity (Ghasemi et al., 2011), indicating digestion of bacteria or antimicrobial protection. The non-reducing disaccharide trehalose forms the primary hemolymph sugar in insects (Shukla et al., 2015) and is cleaved by trehalase to meet energy requirements (Candy and Kilby, 1961; Becker et al., 1996; Reyes-DelaTorre et al., 2012). In millipedes, trehalase activity has been detected in eight of nine millipede species studied (Nielsen, 1962; Marcuzzi and Turchetto Lafisca, 1976, 1978; Nunez and Crawford, 1976; Shukla and Shukla, 1980, 1986; Beck and Friebe, 1981; Shukla, 1984; Šustr et al., 2020a). In A. gigas, trehalase was detected in the contents and in the walls of the midgut and hindgut. The highest activity was detected in the contents of the midgut (Šustr et al., 2020a). Of the CAZy families known to comprise trehalases, only GH15 and GH65 were expressed in the T. aoutii holobiont, both from host tissue (Figure 11B). GH15 is known to contain amylolytic enzymes in addition to trehalases, and its expression in arthropods is not confirmed in CAZy (CAZy, see footnote 1).
Expression of GH65 in the host could indicate an endogenous trehalase, but all transcripts were annotated as glucosylgalactosyl hydroxylysine glucosidase. Moreover, both the GH15 and GH65 families, which belong to the same GH clan-L, include only bacterial and fungal trehalases (Shukla et al., 2015). The GH37 family, known to be universally distributed, including in animals (Shukla et al., 2015), was not expressed in the T. aoutii holobiont. Expression of GH15 and GH65 was distributed in all parts of the millipede, with the highest abundance in the midgut. When considering trehalase in the metatranscriptomic data from the holobiont of T. aoutii, the most striking feature is the different message resulting from the distribution of CAZy and EC numbers. The potentially trehalolytic CAZy families showed the greatest variability in the midgut and were expressed exclusively in the host, whereas a trehalase activity (EC 3.2.1.28) was expressed only by bacteria and fungi in the hindgut (Supplementary Figure S14). The membership of these microbial enzymes in the CAZy family was not established (Table 1, Supplementary Figures S8–S10, and Supplementary Tables S3, S4).
In arthropods, chitin is a structural component of the cuticle, and chitinolytic activity is known to play a non-digestive role during molting. The host in the holobiont of T. aoutii expresses chitinolytic families (GH16, GH18, or GH22) in all body parts. The GH18 family is known for archaea, prokaryotes, and eukaryotes (Funkhouser and Aronson, 2007). The AA15 family includes lytic chitin monooxygenase (EC 1.14.99.53) as one of two EC numbers known in the family. The family is also expressed in tissues other than the intestine of the holobiont T. aoutii. It supports its chitinolytic role in the millipede. Lytic chitin monooxygenase is the only known AA11 family activity. The family is expressed in midgut fungi in the holobiont of T. aoutii. Ciliates in the hindgut express mainly GH25 and to a lesser extent GH22 and GH19. Bacteria exhibited a greater diversity of expressed chitinolytic families, including GH23, GH24, GH25, GH48, and GH73 in addition to GH18 and GH19. Bacteria expressed five potentially chitinolytic families in the midgut and six in the hindgut (Figures 11C,D). In summary, chitinases including CAZy families, and chitinolytic enzymes were expressed by both the host and the gut microbiota, with endogenous expression predominating in the midgut and microbial expression predominating in the hindgut (Supplementary Figure S15).
Most endogenous chitinases (EC 3.2.1.14) present in all host tissues have been identified as GH18 or associated with CBM14. A chitinase belonging to GH19 was expressed in ciliates. Bacterial chitinases were expressed mainly in the hindgut, to a lesser extent in the midgut, and rarely in the foregut. Most of them belonged to GH18 or GH19 and CBMs (5, 6, and 37). The GH18 and GH19 families are known to be widely distributed in the Gammaproteobacteria; GH18 is more commonly reported from the Firmicutes (CAZy, see footnote 1).
Enzymes with lysozyme activity (EC 3.2.1.17), recognized as GH22, were expressed in all host tissues, with similar abundance in millipede midgut and non-gut tissues. Only lysozyme is known from GH22, which has been described in eukaryotes, including arthropods (Lee and Brey, 1995). Ciliates express proteins belonging to the GH25 and GH22 families in the T. aoutii. Only lysozyme activity is known from these two families. Bacterial lysozymes belonging to GH24, GH18, and GH19 families were expressed in the midgut and hindgut. The GH24 family includes only lysozyme activity (CAZy, see footnote 1). Some bacterial activities were further associated with CBM5 and CBM50 in the holobiont of T. aoutii.
The production of β-N-acetylhexosaminidases (EC 3.2.1.52) responsible for the second step of chitin degradation was confirmed both in the host, hindgut ciliates and in the bacterial community (predominantly in the hindgut). The β-N-acetylhexosaminidases expressed in host and hindgut ciliates were identified as GH20 members. The presence of β-N-acetylhexosaminidase in the host confirmed the ability of the millipede to completely degrade chitin by its own enzymatic machinery. The GH20 family is known from bacteria and many eukaryotes, and β-N-acetylhexosaminidase is known from nematodes (Gutternigg et al., 2007) and insects (Kokuho et al., 2010). The β-N-acetylhexosaminidase, belonging to GH20, was expressed in ciliates from the hindgut of T. aoutii, together with chitinases belonging to the GH19 family. The GH19 family contains putative chitinases as well as lysozyme and is found in plants, bacteria, viruses, nematodes, fungi, and other eukaryotic microbes including the supergroup SAR. On the other hand, GH18 has been found in anaerobic ciliates (Park et al., 2021). A complete set of chitinolytic enzymes was found in sheep rumen ciliates (Bełzecki et al., 2008), and chitinases and lysozymes are thought to be essential for protozoan digestion of engulfed fungi and bacteria (Komatani et al., 1997). The bacterial β-N-acetylhexosaminidases in T. aoutii belonged to GH20 and GH3 or were associated with CBM32. Chitosanase activities (EC 3.2.1.132), associated with GH46 were expressed in bacteria in all intestinal sections (Supplementary Tables S3, S4).
Effects of the distribution of carbohydrate-active enzyme in the gut on the energy budget and ecological role of the millipede
The taxonomic composition of saccharolytic enzymes and their distribution among the main intestinal sections is summarized in Figure 12. The midgut of T. aoutii had the greatest diversity of endogenous CAZymes, especially those involved in starch degradation (Figure 13). The large diversity of endogenous amylolytic genes in the midgut may help millipedes to utilize the low levels of storage polysaccharides that enter the system with partially decomposed leaf litter. Millipedes also occasionally feed on a variety of higher-value foods, such as fruits, seeds, fungi, feces, and dead invertebrates (David, 2015); alternatively, the millipede may utilize some of the ingested or intestinal microorganisms (Byzov, 2006) and digest their α-glucans.
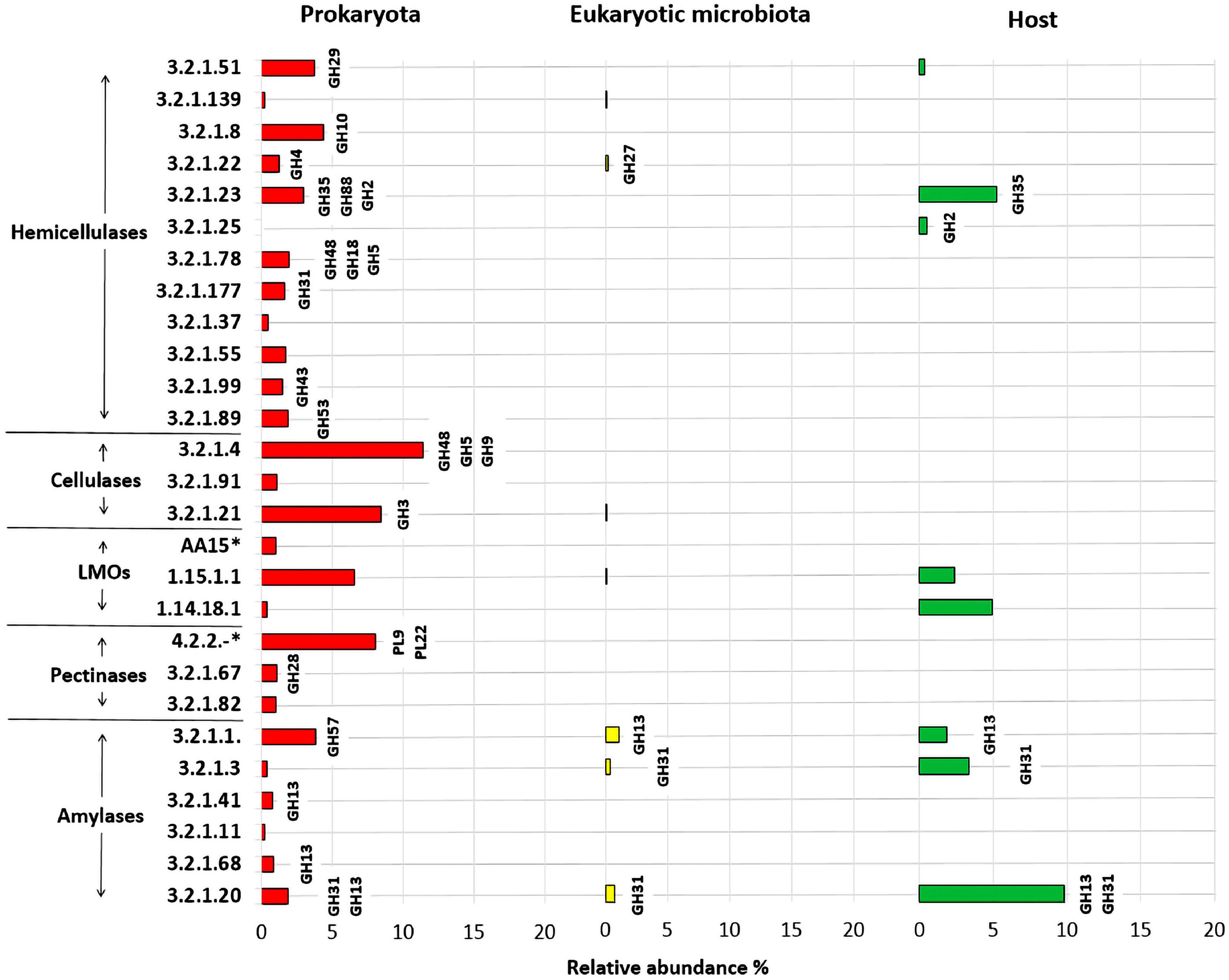
Figure 12. Prediction of enzymatic functions (EC numbers) of hemicellulases, cellulases, pectinases, amylases, lytic polysaccharide monooxygenases, and lignin modifying enzymes (LMOs) expressed in the intestinal prokaryotes, eukaryotic microbiota and the host (T. aoutii). Relative abundance (in%) for a given predicted enzymatic function was calculated by dividing the identified counts for a given enzyme by the total counts identified in the holobiont. 4.2.2.-* – lyases in general, AA15* –EC 1.14.99.54 or EC 1.14.99.53.
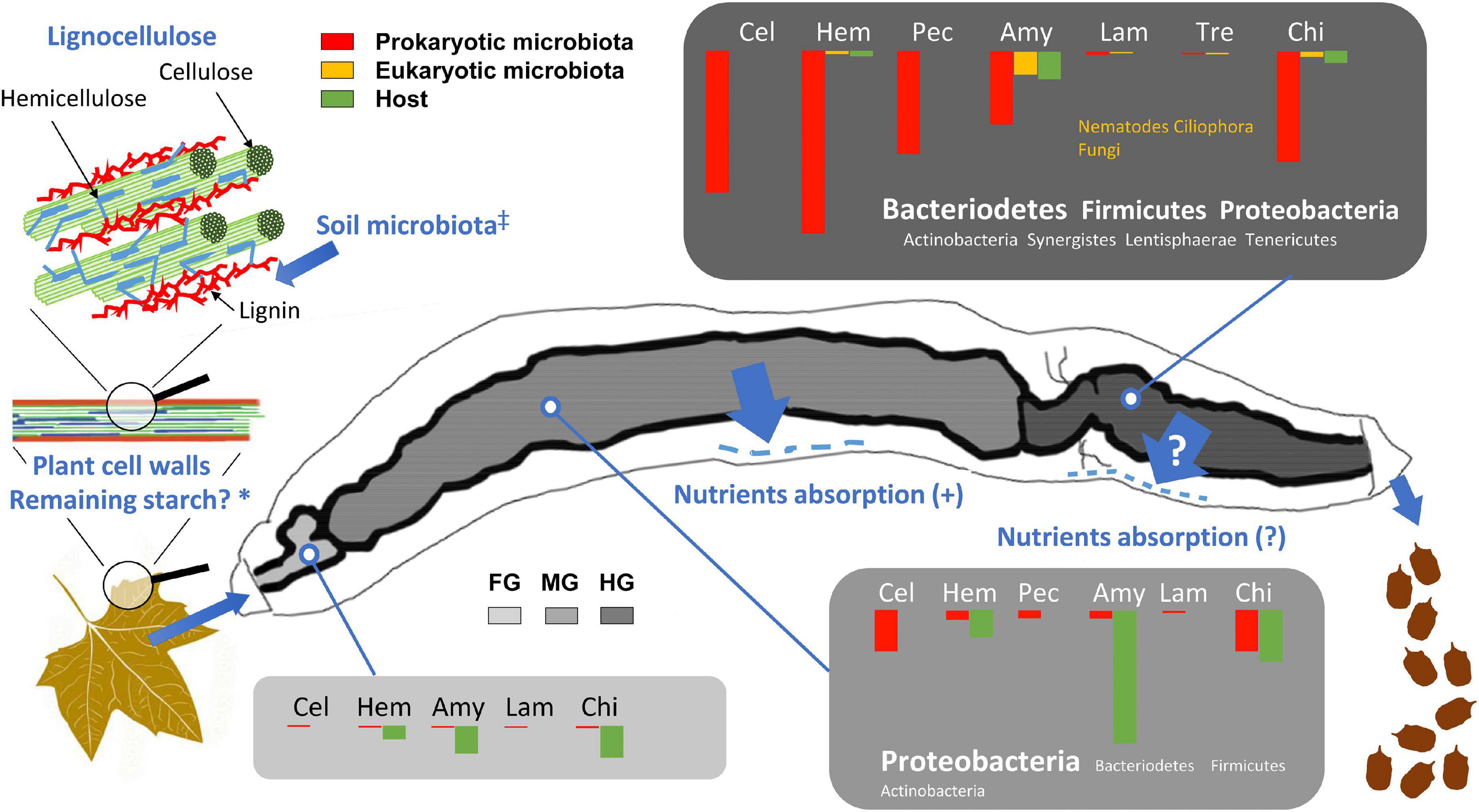
Figure 13. Distribution of expressed CAZymes involved in the digestive processes and degradation of lignocellulose in the gut of the millipede T. aoutii. The length of the bars corresponds to the percentage of the corresponding EC numbers (100% = number of ECs corresponding to all functional groups of CAZymes in the gut). FG, foregut with salivary glands; MG, midgut; HG, hindgut; Cel, cellulases and β-glucosidases; Hem, hemicellulases; Pec, pectinases; Amy, amylases and α-glucosidases; Lam, laminarinases and licheninases; Tre, trehalase; Chi, chitinases and lysozyme. *Residual plant starch or storage polysaccharides of the soil microbiota are expected in the diet, ‡Enzymatic degradation of lignin by the free-living soil microbiota is assumed, Nutrients absorption (?) – Nutrients uptake through the hindgut wall has not been experimentally demonstrated in millipedes. The scheme of lignocellulose was modified after Salimi et al. (2016) and the gut silhouette after Blower (1985).
Although the bacterial community in the midgut expresses some cellulases, the hindgut represents the hotspot of bacterial cellulose degradation (Figure 13). Partial degradation of hemicelluloses, which protect cellulose fibrils in the lignocellulolytic complex, is required as the first step in the digestion of native lignocellulosic material and appears to occur only in the hindgut of the millipede. Degradation of lignin, which has been discussed as a requirement for effective digestion of the lignocellulosic complex (Bredon et al., 2018), has not been documented in T. aoutii. The contribution of the eukaryotic microbiota (nematodes, ciliates, and fungi) to the CAZyme production was less significant. The expression of fungal CAZymes was very rare in the intestine of T. aoutii, which may be due to the anaerobic conditions in most intestinal sections (Horváthová et al., 2021). The expression of amylolytic or chitinolytic activities in nematodes and ciliates, living in the hindgut, supported their commensal feeding on the rich bacterial community.
This spatial distribution of lignocellulose degradation processes in the digestive tract did not indicate effective utilization of energy from lignocellulose degradation by the millipede, as most of the assimilable nutrients are probably already absorbed in the midgut. Thus, the overall importance of lignocellulose digestion to the millipede’s diet depends on the animal’s ability to assimilate microbial metabolites via the hindgut epithelium, which needs to be confirmed by further studies. Nevertheless, previous isotope tracking experiments with 14C-labeled cellulose in several millipede species (Taylor, 1982; Bignell, 1989) confirmed the assimilation of cellulose by millipedes and the role of microorganisms in the digestive process.
The low contribution of energy derived from the digestion of lignocellulose to the energy budget of millipedes can be inferred from the high potential of endogenous amylolytic activity as evidenced by metatranscriptomic and enzymological data, the low assimilation efficiency of millipedes (Hopkin and Read, 1992), and the low metabolic rate of millipedes fed pure cellulose (Šustr et al., 2020b). This suggests a preferential intake of more easily digestible dietary energy sources. The millipede Glomeris marginata assimilated simple sugars with an efficiency of 100% and amino acids with an efficiency of up to 95%, whereas the fibers of beech leaves were assimilated at only 18% (Bignell, 1989). In addition, millipedes feed on decaying plant material colonized by microbial decomposers (bacteria and fungi) (David, 2015), and incomplete digestion of lignocellulose in the midgut can be partially compensated by consumption of food partially degraded by the soil microbiota and by mechanical fragmentation of food by the mouthparts. It is possible that the soil microbiota may partly degrade lignin before it is ingested by the millipede.
If the millipede did not utilize the products of hindgut microbial activity, the hindgut microbiota would rather be commensals, and its enzyme activity would be of no nutritional significance to the host. On the other hand, the activity of the hindgut microbiota may be important for the ecological role of the millipede as a holobiont. Due to the low assimilation efficiency of millipedes, a large amount of fecal material, modified by the activity of the hindgut microbiota is deposited in the soil by the millipede. The deposition of fecal pellets modified by the activity of hindgut microbiota represents an important ecological service of the holobiont.
Conclusion
Here we have provided some insight into the digestion and microbial breakdown of carbohydrates in the millipede species T. aoutii. In general, easily digestible substrates are likely broken down endogenously in the midgut, as indicated by the large diversity of endogenous amylases and α-glucosidases in the midgut (Figure 13). These enzymes effectively break down α-glucans found in the millipede diet as residual plant storage polysaccharides in partially decomposed leaf litter, ingested microorganisms, and other occasional food sources. These α-polysaccharides can provide an important source of energy and carbon for millipedes and the products of their cleavage can be readily absorbed by the midgut epithelium. In contrast, no transcripts of endogenous cellulases were detected, and the cellulolytic and hemicellulolytic potential of the midgut microbial community is limited. Genes encoding cellulases and hemicellulases are transcribed primarily by the prokaryotic microbiota of the hindgut (bacteria) (Figure 13). Most microbial transcripts containing CAZymes belonged to bacteria: CAZyme families expressed by Proteobacteria generally predominate in the gut, whereas families produced by Bacteroidetes and Firmicutes are abundant in the hindgut. The contribution of intestinal fungi and other eukaryotic microbiota to CAZymes production was negligible. Overall, the enzymatic apparatus of the holobiont is probably capable of degrading cellulose, hemicelluloses, starch, laminarin or lichenin, sucrose, trehalose, and chitin, but not lignin. The breakdown of the lignin-impregnated parts of millipede food most likely depends on their partial degradation by ligninolytic microorganisms in the soil litter horizon prior to their consumption. The extent to which the relationship between the cellulolytic gut bacteria provides important benefits to the millipede’s ability of the millipede to absorb and utilize microbial metabolites as nutrients through the hindgut wall which needs to be demonstrated by further studies. In summary, T. aoutii can be considered a decomposer of leaf litter and soil organic matter depending primarily on its bacterial community in the hindgut. The efficiency of the food assimilation in millipedes is usually not high and a large amount of food must be ingested to meet metabolic needs. The resulting large production of fecal pellets, influenced by the hindgut microbiota of millipedes, could be of great ecological importance for the transformation of soil organic matter.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA749320.
Author contributions
PS designed and performed the all bioinformatics and wrote the relevant section of the manuscript. VŠ wrote the first draft of the manuscript. VŠ, PS, and AC contributed to conception and design of the study. VŠ and FL contributed to evaluation and interpretation of functional annotation outputs. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported by the Czech Science Foundation (GACR) (project No. 17-22572S). PS was supported by European Structural and Investment Funds, project MEMOBiC (No. 551 CZ.02.2.69/0.0/0.0/16_027/0008357).
Acknowledgments
The authors thank M. Šimek for comments to the manuscript, L. Faktorová, M. Petrlíková, and A. Koubová for assistance in laboratory analyses and Š. Otáhalová for improving tables and figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.931986/full#supplementary-material
Footnotes
References
Albersheim, P., Darvill, A., Roberts, K., Sederoff, R., and Staehelin, A. (2011). Plant cell walls: From chemistry to biology. New York, NY: Garland Science. doi: 10.1201/9780203833476
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bacic, A., Fincher, G. B., and Stone, B. A. (2009). in Chemistry, biochemistry, and biology of 1-3 beta glucans and related polysaccharides, eds A. Bacic, G. B. Fincher, and B. A. Stone San Diego (Cambridge, MA: Academic Press). doi: 10.1016/B978-0-12-373971-1.00025-X
Beck, L., and Friebe, B. (1981). Verwertung von Kohlenhydraten bei Oniscus asellus (Isopoda) und Polydesmus angustus (Diplopoda). Pedobiologia 21, 19–29.
Becker, A., Schlöder, P., Steele, J. E., and Wegener, G. (1996). The regulation of trehalose metabolism in insects. Experientia 52, 433–439. doi: 10.1007/BF01919312
Bełzecki, G., Miltko, R., Michałowski, T., Šimůnek, J., and Kopečný, J. (2008). Chitinolytic activity of the sheep rumen ciliate Diploplastron affine. Folia Microbiol. 53:201. doi: 10.1007/s12223-008-0025-y
Berg, M. P., Stoffer, M., and van den Heuvel, H. H. (2004). Feeding guilds in Collembola based on digestive enzymes. Pedobiologia 48, 589–601. doi: 10.1016/j.pedobi.2004.07.006
Besser, K., Malyon, G. P., Eborall, W. S., Paro da Cunha, G., Filgueiras, J. G., Dowle, A., et al. (2018). Hemocyanin facilitates lignocellulose digestion by wood-boring marine crustaceans. Nat. Commun. 9:5125. doi: 10.1038/s41467-018-07575-2
Bignell, D. E. (1989). Relative assimilations of 14C-labelled microbial tissues and 14C-plant fibre ingested with leaf litter by the millipede Glomeris marginata under experimental conditions. Soil Biol. Biochem. 21, 819–827. doi: 10.1016/0038-0717(89)90176-4
Blesák, K., and Janeček, S. (2012). Sequence fingerprints of enzyme specificities from the glycoside hydrolase family GH57. Extremophiles 16, 497–506. doi: 10.1007/s00792-012-0449-9
Blower, J. G. (1985). “Millipedes,” in Synopsis of the British Fauna (NS 35, (London: EJ: Backhuys Publishers).
Bogdanova, E. A., Shagina, I. A., Ianushevich, I. G., Vagner, L. L., Luk’ianov, S. A., and Shagin, D. A. (2011). Preparation of prokaryotic cDNA for high-throughput transcriptome analysis. Bioorg. Khim. 37, 854–857. doi: 10.1134/s1068162011060045
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bredon, M., Dittmer, J., Noel, C., Moumen, B., and Bouchon, D. (2018). Lignocellulose degradation at the holobiont level: Teamwork in a keystone soil invertebrate. Microbiome 6:162. doi: 10.1186/s40168-018-0536-y
Bredon, M., Herran, B., Bertaux, J., Grève, P., Moumen, B., and Bouchon, D. (2020). Isopod holobionts as promising models for lignocellulose degradation. Biotechnol. Biofuels 13:49. doi: 10.1186/s13068-020-01683-2
Brune, A., and Ohkuma, M. (2011). “Role of the Termite Gut Microbiota in Symbiotic Digestion,” in Biology of termites: A modern synthesis, eds D. E. Bignell, Y. Roisin, and N. Lo (Dordrecht: Springer Netherlands), 439–475. doi: 10.1007/978-90-481-3977-4_16
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Busk, P. K., Busk, P. K., Lezyk, M. J., Meyer, A. S., and Lange, L. (2017). Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinform. 18:214. doi: 10.1186/s12859-017-1625-9
Byeon, G. M., Lee, K. S., Gui, Z. Z., Kim, I., Kang, P. D., Lee, S. M., et al. (2005). A digestive beta-glucosidase from the silkworm, Bombyx mori: cDNA cloning, expression and enzymatic characterization. Comparative biochemistry and physiology. Part B, Biochem. Mol. Biol. 141, 418–427. doi: 10.1016/j.cbpc.2005.05.001
Byzov, B. A. (2006). “Intestinal Microbiota of Millipedes,” in Intestinal microorganisms of termites and other invertebrates, (Berlin: Springer), 89–114. doi: 10.1007/3-540-28185-1_4
Candy, D. J., and Kilby, B. A. (1961). The biosynthesis of trehalose in the locust fat body. Biochem. J. 78, 531–536. doi: 10.1042/bj0780531
Cazemier, A. E., Den Camp, H. J. M. O., Hackstein, J. H. P., and Vogels, G. D. (1997). Fibre digestion in arthropods. Comp. Biochem. Physiol. A Physiol. 118, 101–109. doi: 10.1016/S0300-9629(96)00443-4
Ceja-Navarro, J. A., Karaoz, U., Bill, M., Hao, Z., White, R. A., Arellano, A., et al. (2019). Gut anatomical properties and microbial functional assembly promote lignocellulose deconstruction and colony subsistence of a wood-feeding beetle. Nat. Microbiol. 4, 864–875. doi: 10.1038/s41564-019-0384-y
Cragg, S., Beckham, G., Bruce, N., Bugg, T., Distel, D., Dupree, P., et al. (2015). Lignocellulose degradation mechanisms across the Tree of Life. Curr. Opin Chem. Biol. 29, 108–119. doi: 10.1016/j.cbpa.2015.10.018
da Costa-Latgé, S. G., Bates, P., Dillon, R., and Genta, F. A. (2021). Characterization of glycoside hydrolase families 13 and 31 reveals expansion and diversification of α-amylase genes in the Phlebotomine Lutzomyia longipalpis and modulation of sandfly glycosidase activities by Leishmania Infection. Front. Physiol. 12:635633. doi: 10.3389/fphys.2021.635633
Da Lage, J.-L. (2018). The amylases of insects. Int. J. Insect Sci. 10:1179543318804783. doi: 10.1177/1179543318804783
Da Lage, J.-L., Danchin, E. G. J., and Casane, D. (2007). Where do animal alpha-amylases come from? An interkingdom trip. FEBS Lett. 581, 3927–3935. doi: 10.1016/j.febslet.2007.07.019
Damsgaard, C., Fago, A., Hagner-Holler, S., Malte, H., Burmester, T., and Weber, R. E. (2013). Molecular and functional characterization of hemocyanin of the giant African millipede, Archispirostreptus gigas. J. Exp. Biol. 216, 1616–1623. doi: 10.1242/jeb.080861
David, J. F. (2014). The role of litter-feeding macroarthropods in decomposition processes: A reappraisal of common views. Soil Biol. Biochem. 76, 109–118. doi: 10.1016/j.soilbio.2014.05.009
David, J.-F. (2015). Diplopoda – Ecology, in Treatise on zoology – Anatomy, taxonomy, biology. The myriapoda, Vol. ed. A. Minelli (Leiden: Brill), 2.
Dedeine, F., Weinert, L. A., Bigot, D., Josse, T., Ballenghien, M., Cahais, V., et al. (2015). Comparative analysis of transcriptomes from secondary reproductives of three Reticulitermes termite species. PLoS One 10:e0145596. doi: 10.1371/journal.pone.0145596
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Erban, T., and Hubert, J. (2008). Digestive function of lysozyme in synanthropic acaridid mites enables utilization of bacteria as a food source. Exp. Appl. Acarol. 44:199. doi: 10.1007/s10493-008-9138-x
Ezeilo, U. R., Zakaria, I. I., Huyop, F., and Wahab, R. A. (2017). Enzymatic breakdown of lignocellulosic biomass: The role of glycosyl hydrolases and lytic polysaccharide monooxygenases. Biotechnol. Biotechnol. Equip. 31, 647–662. doi: 10.1080/13102818.2017.1330124
Fontanetti, C. S., Moreira-de-Sousa, C., Pinheiro, T. G., de Souza, R. B., and Francisco, A. (2015). “Diplopoda — digestive system,” in Treatise on zoology–Anatomy, taxonomy, biology. The myriapoda, ed. A. Minelli (Leiden: Brill), 2. doi: 10.1163/9789004188273_006
Funkhouser, J. D., and Aronson, N. N. (2007). Chitinase family GH18: Evolutionary insights from the genomic history of a diverse protein family. BMC Evol. Biol. 7:96. doi: 10.1186/1471-2148-7-96
Gabriško, M. (2013). Evolutionary history of eukaryotic α-glucosidases from the α-amylase family. J. Mol. Evol. 76, 129–145. doi: 10.1007/s00239-013-9545-4
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Geng, A., Jin, M., Li, N., Zhu, D., Xie, R., Wang, Q., et al. (2021). New insights into the co-occurrences of glycoside hydrolase genes among prokaryotic genomes through network analysis. Microorganisms 9:427. doi: 10.3390/microorganisms9020427
Ghasemi, S., Ahmadian, G., Sadeghi, M., Zeigler, D. R., Rahimian, H., Ghandili, S., et al. (2011). First report of a bifunctional chitinase/lysozyme produced by Bacillus pumilus SG2. Enzyme Microb. Technol. 48, 225–231. doi: 10.1016/j.enzmictec.2010.11.001
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Griffiths, H. M., Ashton, L. A., Parr, C. L., and Eggleton, P. (2021). The impact of invertebrate decomposers on plants and soil. New Phytol. 231, 2142–2149.
Gutternigg, M., Kretschmer-Lubich, D., Paschinger, K., Rendić, D., Hader, J., Geier, P., et al. (2007). Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants, and insects. J. Biol. Chem. 282, 27825–27840. doi: 10.1074/jbc.M704235200
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
He, S., Ivanova, N., Kirton, E., Allgaier, M., Bergin, C., Scheffrahn, R. H., et al. (2013). Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8:e61126. doi: 10.1371/journal.pone.0061126
Hii, S. L., Tan, J. S., Ling, T. C., and Ariff, A. B. (2012). Pullulanase: Role in starch hydrolysis and potential industrial applications. Enzyme Res. 2012:921362. doi: 10.1155/2012/921362
Horváthová, T., Šustr, V., Chroňáková, A., Semanová, S., Lang, K., Dietrich, C., et al. (2021). Methanogenesis in the digestive tracts of the tropical millipedes Archispirostreptus gigas (Diplopoda, Spirostreptidae) and Epibolus pulchripes (Diplopoda, Pachybolidae). Appl. Environ. Microbiol. 87:e0061421. doi: 10.1128/AEM.00614-21
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Huson, D. H., Beier, S., Flade, I., Górska, A., El-Hadidi, M., Mitra, S., et al. (2016). MEGAN Community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comp. Biol. 12:e1004957. doi: 10.1371/journal.pcbi.1004957
Ishida, Y., Kuwahara, Y., Dadashipour, M., Ina, A., Yamaguchi, T., Morita, M., et al. (2016). A sacrificial millipede altruistically protects its swarm using a drone blood enzyme, mandelonitrile oxidase. Sci. Rep. 6:26998. doi: 10.1038/srep26998
Janeček, Š, and Svensson, B. (2022). How many α-amylase GH families are there in the CAZy database? Amylase 6, 1–10. doi: 10.1515/amylase-2022-0001
Jorge, J. A., Polizeli, M. L., Thevelein, J. M., and Terenzi, H. F. (1997). Trehalases and trehalose hydrolysis in fungi. FEMS Microbiol. Lett. 154, 165–171. doi: 10.1111/j.1574-6968.1997.tb12639.x
Kane, M. D. (1997). “Microbial Fermentation in Insect Guts,” in Gastrointestinal microbiology: Volume 1 Gastrointestinal ecosystems and fermentations, eds R. I. Mackie and B. A. White (Boston, MA: Springer US), 231–265. doi: 10.1007/978-1-4615-4111-0_8
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kellokumpu, S., Hassinen, A., and Glumoff, T. (2016). Glycosyltransferase complexes in eukaryotes: Long-known, prevalent but still unrecognized. Cell. Mol. Life Sci. 73, 305–325. doi: 10.1007/s00018-015-2066-0
Kohn, R., and Luknár, O. (1977). Intermolecular calcium ion binding on polyuronates-polygalacturonate and polyguluronate. Collect. Czechoslov. Chem. Commun. 42:731. doi: 10.1135/cccc19770731
Kokuho, T., Yasukochi, Y., Watanabe, S., and Inumaru, S. (2010). Molecular cloning and expression profile analysis of a novel beta-D-N-acetylhexosaminidase of domestic silkworm (Bombyx mori). Genes to cells 15, 525–536. doi: 10.1111/j.1365-2443.2010.01401.x
Komatani, K., Morgavi, D. P., Onodera, R., White, B. A., Cann, I., and Karrer, K. M. (1997). Molecular cloning of a chitinase gene from the rumen protozoon Entodinium caudatum. EDP Sci. 37, 48–49.
Kopylova, E., Noé, L., and Touzet, H. (2012). SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lee, W. J., and Brey, P. T. (1995). Isolation and characterization of the lysozyme-encoding gene from the silkworm Bombyx mori. Gene 161, 199–203. doi: 10.1016/0378-1119(95)00199-g
Li, W., and Godzik, A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lo, N., Tokuda, G., and Watanabe, H. (2011). “Evolution and function of endogenous termite cellulases,” in Biology of termites: A modern synthesis, eds D. E. Bignell, Y. Roisin, and N. Lo (Dordrecht: Springer Netherlands), 51–67. doi: 10.1007/978-90-481-3977-4_3
Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleid Acid Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Luo, C., Li, Y., Liao, H., and Yang, Y. (2018). De novo transcriptome assembly of the bamboo snout beetle Cyrtotrachelus buqueti reveals ability to degrade lignocellulose of bamboo feedstock. Biotechnol. Biofuels 11:292. doi: 10.1186/s13068-018-1291-9
Marcuzzi, G., and Turchetto Lafisca, M. (1976). Contribute to the knowledge of the digestive enzymes of some litter feeding animals. Oligosaccharases. Revue D Ecologie et de Biologie du Sol 13, 449–458.
Marcuzzi, G., and Turchetto Lafisca, M. (1978). Contribute to the knowledge of polysaccharases in soil animals. Revue D Ecologie et de Biologie du Sol 15, 135–145.
Matsuura, K., Yashiro, T., Shimizu, K., Tatsumi, S., and Tamura, T. (2009). Cuckoo fungus mimics termite eggs by producing the cellulose-digesting enzyme β-glucosidase. Curr. Biol. 19, 30–36. doi: 10.1016/j.cub.2008.11.030
Melov, S., Ravenscroft, J., Malik, S., Gill, M. S., Walker, D. W., Clayton, P. E., et al. (2000). Extension of life-span with superoxide dismutase/catalase mimetics. Science 289, 1567–1569.
Mills, K. L., Hart, B. J., Lynch, N. R., Thomas, W. R., and Smith, W. (1999). Molecular characterization of the group 4 house dust mite allergen from Dermatophagoides pteronyssinus and its amylase homologue from Euroglyphus maynei. Int. Arch. Allergy Immunol. 120, 100–107. doi: 10.1159/000024227
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. doi: 10.1093/nar/gkt263
Ni, J., and Tokuda, G. (2013). Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Advan. 31, 838–850. doi: 10.1016/j.biotechadv.2013.04.005
Nielsen, C. (1962). Carbohydrases in soil and litter invertebrates. Oikos 13, 200–215. doi: 10.2307/3565085
Nunez, F. S., and Crawford, C. S. (1976). Digestive enzymes of the desert millipede Orthoporus ornatus (Girard) (Diplopoda: Spirostreptidae). Comp. Biochem. Physiol. Part A Physiol. 55, 141–145. doi: 10.1016/0300-9629(76)90082-7
Ohashi, K., Sawata, M., Takeuchi, H., Natori, S., and Kubo, T. (1996). Molecular cloning of cDNA and analysis of expression of the gene for alpha-glucosidase from the hypopharyngeal gland of the honeybee Apis mellifera L. Biochem. Biophys. Res. Commun. 221, 380–385. doi: 10.1006/bbrc.1996.0604
Olianas, A., Sanjust, E., Pellegrini, M., and Rescigno, A. (2005). Tyrosinase activity and hemocyanin in the hemolymph of the slipper lobster Scyllarides latus. J. Comp. Physiol. B 175, 405–411. doi: 10.1007/s00360-005-0002-6
Ordaz-Ortiz, J. J., and Saulnier, L. (2005). Structural variability of arabinoxylans from wheat flour. Comparison of water-extractable and xylanase-extractable arabinoxylans. J. Cereal Sci. 42, 119–125. doi: 10.1016/j.jcs.2005.02.004
Palomares-Rius, J. E., Hirooka, Y., Tsai, I. J., Masuya, H., Hino, A., Kanzaki, N., et al. (2014). Distribution and evolution of glycoside hydrolase family 45 cellulases in nematodes and fungi. BMC Evol. Biol. 14:69. doi: 10.1186/1471-2148-14-69
Park, T., Wijeratne, S., Meulia, T., Firkins, J. L., and Yu, Z. (2021). The macronuclear genome of anaerobic ciliate Entodinium caudatum reveals its biological features adapted to the distinct rumen environment. Genomics 113, 1416–1427. doi: 10.1016/j.ygeno.2021.03.014
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Piavaux, A. (1977). Distribution and localization of the digestive laminarinases in animals. Biochem. Syst. Ecol. 5, 231–239.
R Core Team. (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna: R Core Team.
Rashid, G. M. M., Taylor, C. R., Liu, Y., Zhang, X., Rea, D., Fülöp, V., et al. (2015). Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation. ACS Chem. Biol. 10, 2286–2294. doi: 10.1021/acschembio.5b00298
Reyes-DelaTorre, A., Pena-Rangel, M. T., and Riesgo-Escovar, J. R. (2012). “Carbohydrate metabolism in Drosophila: Reliance on the disaccharide trehalose,” in Carbohydrates, ed. C. F. Chang (Rijeka: IntechOpen). doi: 10.5772/50633
Sabbadin, F., Hemsworth, G. R., Ciano, L., Henrissat, B., Dupree, P., Tryfona, T., et al. (2018). An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 9:756. doi: 10.1038/s41467-018-03142-x
Sakamoto, Y., Watanabe, H., Nagai, M., Nakade, K., Takahashi, M., and Sato, T. (2006). Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 141, 793–801. doi: 10.1104/pp.106.076679
Salimi, M., Nejati, B., Karimi, A., and Tavasoli, A. (2016). Hydrothermal gasification performance of iranian rice straw in supercritical water media for hydrogen-rich gas production. Bioresources 11, 6362–6377.
Scharf, M. E. (2015). Termites as targets and models for biotechnology. Annu. Rev. Entomol. 60, 77–102. doi: 10.1146/annurev-ento-010814-020902
Scully, E. D., Geib, S. M., Carlson, J. E., Tien, M., McKenna, D., and Hoover, K. (2014). Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics 15:1096. doi: 10.1186/1471-2164-15-1096
Shelomi, M., Danchin, E. G. J., Heckel, D., Wipfler, B., Bradler, S., Zhou, X., et al. (2016). Horizontal gene transfer of pectinases from bacteria preceded the diversification of stick and leaf insects. Sci. Rep. 6:26388. doi: 10.1038/srep26388
Shimada, K., and Maekawa, K. (2014). Gene expression and molecular phylogenetic analyses of beta-glucosidase in the termite Reticulitermes speratus (Isoptera: Rhinotermitidae). J. Insect Physiol. 65, 63–69. doi: 10.1016/j.jinsphys.2014.05.006
Shukla, E., Thorat, L. J., Nath, B. B., and Gaikwad, S. M. (2015). Insect trehalase: Physiological significance and potential applications. Glycobiology 25, 357–367. doi: 10.1093/glycob/cwu125
Shukla, G. S., and Shukla, S. C. (1980). Nature of gut invertase in millipede Trigoniulus lumbricinus (Gerstacker) with special reference to its activity in the midgut. J. Adv. Zool. 1, 86–93.
Shukla, G. S., and Shukla, S. C. (1986). Physiology of digestion in the millipede Trigoniulus lumbricinus (Gerstacker). Indian J. Entomol. 48, 281–288.
Shukla, S. C. (1984). Trehalase activity in the millipede Trigoniulus lumbricinus (Gerstacker). Comp. Physiol. Ecol. 9, 56–58.
Siepel, H., and de Ruiter-Dijkman, E. M. (1993). Feeding guilds of oribatid mites based on their carbohydrase activities. Soil Biol. Biochem. 25, 1491–1497. doi: 10.1016/0038-0717(93)90004-U
Singh, G., Verma, A. K., and Kumar, V. (2016). Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech 6:3. doi: 10.1007/s13205-015-0328-z
Šustr, V., Semanová, S., Rost-Roszkowska, M. M., Tajovskı, K., Sosinka, A., and Kaszuba, F. (2020a). Enzymatic activities in the digestive tract of spirostreptid and spirobolid millipedes (Diplopoda: Spirostreptida and Spirobolida). Comparative biochemistry and physiology. Part B, Biochem. Mol. Biol. 241:110388. doi: 10.1016/j.cbpb.2019.110388
Šustr, V., Šimek, M., Faktorová, L., Macková, J., and Tajovský, K. (2020b). Release of greenhouse gases from millipedes as related to food, body size, and other factors. Soil Biol. Biochem. 144:107765. doi: 10.1016/j.soilbio.2020.107765
Taylor, E. C. (1982). Role of aerobic microbial populations in cellulose digestion by desert millipedes. Appl. Environ. Microbiol. 44, 281–291. doi: 10.1128/aem.44.2.281-291.1982
Terra, W. R., and Ferreira, C. (1994). Insect digestive enzymes: Properties, compartmentalization and function. Comp. Biochem. Physiol. Part B Comp. Biochem. 109, 1–62. doi: 10.1016/0305-0491(94)90141-4
Terra, W. R., and Ferreira, C. (2005). “4.5 - Biochemistry of digestion,” in Comprehensive molecular insect science, ed. L. I. Gilbert (Amsterdam: Elsevier), 171–224. doi: 10.1016/B0-44-451924-6/00053-3
Tokuda, G. (2019). “Chapter Three - Plant cell wall degradation in insects: Recent progress on endogenous enzymes revealed by multi-omics technologies,” in Advances in insect physiology, ed. R. Jurenka (Cambridge, MA: Academic Press), 97–136. doi: 10.1016/bs.aiip.2019.08.001
Tokuda, G., Lo, N., and Watanabe, H. (2005). Marked variations in patterns of cellulase activity against crystalline- vs. carboxymethyl-cellulose in the digestive systems of diverse, wood-feeding termites. Physiol. Entomol. 30, 372–380. doi: 10.1111/j.1365-3032.2005.00473.x
Ulaganathan, K., Goud, B. S., Reddy, M. M., Kumar, V. P., Balsingh, J., and Radhakrishna, S. (2015). Proteins for breaking barriers in lignocellulosic bioethanol production. Curr. Protein pept. Sci. 16, 100–134. doi: 10.2174/138920371602150215165718
Urbášek, F., and Tajovský, K. (1991). The influence of food and temperature on enzymatic activities of the millipede Glomeris hexasticha (Diplopoda). Revue D Ecologie et de Biologie du Sol 28, 155–163.
Van der Drift, J. (1951). Analysis of the animal community in a beech forest floor. Tijdschr. Entomol. 94, 1–168.
Venn, J. (1880). On the employment of geometrical diagrams for the sensible representation of logical propositions. Proc. Camb. Philos. Soc. 4, 47–59.
Wagner, G. P., Kin, K., and Lynch, V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285. doi: 10.1007/s12064-012-0162-3
Watanabe, H., and Tokuda, G. (2001). Animal cellulases. Cell. Mol. Life Sci. 58, 1167–1178. doi: 10.1007/PL00000931
Watanabe, H., and Tokuda, G. (2010). Cellulolytic systems in insects. Annu. Rev. Entomol. 55, 609–632. doi: 10.1146/annurev-ento-112408-085319
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., and Xu, Y. (2012). dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451. doi: 10.1093/nar/gks479
Yuki, M., Moriya, S., Inoue, T., and Kudo, T. (2008). Transcriptome analysis of the digestive organs of Hodotermopsis sjostedti, a lower termite that hosts mutualistic microorganisms in its hindgut. Zool. Sci. 25, 401–406. doi: 10.2108/zsj.25.401
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Zhang, R., and Williams, S. (2021). Glycoside hydrolase family 31. CAZypedia. Available online at: https://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_31 (accessed December 7, 2021).
Keywords: Diplopoda, millipede, metatranscriptome, carbohydrate-active enzymes, lignocellulose, holobiont
Citation: Sardar P, Šustr V, Chroňáková A and Lorenc F (2022) Metatranscriptomic holobiont analysis of carbohydrate-active enzymes in the millipede Telodeinopus aoutii (Diplopoda, Spirostreptida). Front. Ecol. Evol. 10:931986. doi: 10.3389/fevo.2022.931986
Received: 29 April 2022; Accepted: 16 August 2022;
Published: 16 September 2022.
Edited by:
Ulf Bauchinger, Jagiellonian University, PolandReviewed by:
Bredon Marius, Sorbonne Université, FranceLingling Ma, Northwest A&F University, China
Yuanqiu Li, Leshan Normal University, China
Copyright © 2022 Sardar, Šustr, Chroňáková and Lorenc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vladimír Šustr, sustr@upb.cas.cz
†Present address: Puspendu Sardar, Cambridge Institute of Therapeutic Immunology and Infectious Disease, Department of Medicine, University of Cambridge, Cambridge, United Kingdom
‡These authors have contributed equally to this work
 Puspendu Sardar
Puspendu Sardar Vladimír Šustr
Vladimír Šustr Alica Chroňáková
Alica Chroňáková František Lorenc
František Lorenc