Abstract
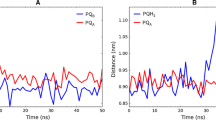
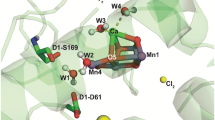
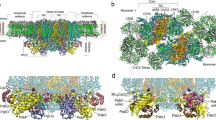
Photosystem II (PSII) is a multi-subunit pigment-protein complex and is one of several protein assemblies that function cooperatively in photosynthesis in plants and cyanobacteria. As more structural data on PSII become available, new questions arise concerning the nature of the charge separation in PSII reaction center (RC). The crystal structure of PSII RC from cyanobacteria Thermosynechococcus vulcanus was selected for the computational study of conformational changes in photosystem II associated to the charge separation process. The parameterization of cofactors and lipids for classical MD simulation with Amber force field was performed. The parametrized complex of PSII was embedded in the lipid membrane for MD simulation with Amber in Gromacs. The conformational behavior of protein and the cofactors directly involved in the charge separation were studied by MD simulations and QM/MM calculations. This study identified the most likely mechanism of the proton-coupled reduction of plastoquinone QB. After the charge separation and the first electron transfer to QB, the system undergoes conformational change allowing the first proton transfer to QB− mediated via Ser264. After the second electron transfer to QBH, the system again adopts conformation allowing the second proton transfer to QBH−. The reduced QBH2 would then leave the binding pocket.









Similar content being viewed by others
Abbreviations
- PQ:
-
Plastoquinone
- PSII:
-
Photosystem II
- Pheo:
-
Pheophytin
- POPC:
-
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- Chl:
-
Chlorophyll
- RC:
-
Reaction center
- Cyt:
-
Cytochrome
- QA :
-
Plastoquinone A
- QB :
-
Plastoquionone B
- DGDG:
-
Digalactosyl-diacylglycerol
- SQDG:
-
Sulfoquinovosyl-diacylglycerol
- MGDG:
-
Monogalactosyl-diacylglycerol
- PG:
-
1,2-Dipalmitoyl-phosphatidyl-glycerol
- uQ:
-
Ubiquinone
- bRC:
-
Bacterial reaction center
References
Vermaas W (1998) An introduction to photosynthesis and its applications The World & I: 158–165
Wydrzynski T, Satoh S (2005) Photosystem II: the light-driven water: plastoquinone oxidoreductase, Springer, Dordrecht
Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W (2009) Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol 16:334–342
Müh F, Glöckner C, Hellmich J, Zouni A (2012) Light-induced quinone reduction in photosystem II. Biochim Biophys Acta 1817:44–65
Zheleva D, Hankamer B, Barber J (1996) Heterogeneity and pigment composition of isolated photosystem II reaction centers. Biochemistry 35:15074–15079
Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0A resolution structure of photosystem II. Nature 438:1040–1044
Biesiadka J, Loll B, Kern J, Irrgang KD, Zouni A (2004) Crystal structure of cyanobacterial photosystem II at 3.2 angstrom resolution: a closer look at the Mn-cluster. Phys Chem Chem Phys 6:4733–4736
Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture ofthephotosyntheticoxygen-evolvingcenter. Science 303:1831–1838
Kamiya N, Shen JR (2003) Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution. Proc Natl Acad Sci U S A 100(1):98–103
Zouni A, Witt H-T, Kern J, Fromme P, Krauss N, Saenger W, Orth P (2001) Crystal structure of photosystem II from Synechococcus elongates at 3.8 Å resolution. Nature 409:739–743
Tanaka A, Fukushima Y, Kamiya N (2017) Two different structures of the oxygen-evolving complex in the same polypeptide frameworks of photosystem II. J Am Chem Soc 139(5):1718–1721
Koua FHM, Umena Y, Kawakami K, Shen J-R (2013) Structure of Sr-substituted photosystem II at 2.1 Å resolution and its implications in the mechanism of water oxidation. Proc Natl Acad Sci U S A 110:3889–3894
Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473:55–60
Wei X, Su X, Cao P, Liu X, Chang W, Li M, Zhang X, Liu Z (2016) Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 534:69–74
Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2007) Lipids in photosystem II: interactions with protein and cofactors. Biochim Biophys Acta 1767:509–519
Van Eerden FJ, Melo MN, Frederix PWJM, Periole X, Marrink SJ (2017) Exchange pathways of plastoquinone and plastoquinol in the photosystem II complex. Nat Commun 8:15214
Okamura MY, Paddock ML, Graige MS, Feher G (2000) Proton and electron transfer in bacterial reaction centers. Biochim Biophys Acta 1458:148–163
Ago H, Adachi H, Umena Y, Tashiro T, Kawakami K, Kamiya N, Tian L, Han G, Kuang T, Liu Z, Wang F, Zou H, Enami I, Miyano M, Shen JR (2016) Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J Biol Chem 291:5676–5687
Krieger E, Koraimann G, Vriend G (2002) Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins 47:393–402
Zayats V, Stockner T, Pandey SK, Wörz K, Ettrich R, Ludwig J (2015) A refined atomic scale model of the Saccharomyces cerevisiae K(+)-translocation protein Trk1p combined with experimental evidence confirms the role of selectivity filter glycines and other key residues. Biochim Biophys Acta 1848:1183–1195
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N•log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Humphrey W, Dalke A, Schulten K (1996) VMD - visual molecular dynamics. J Molec Graphics 14:33–38
Zhang L, Silva DA, Yan Y, Huang X (2012) Force field development for cofactors in the photosystem II. J Comput Chem 33:1969–1980
Jämbeck JPM, Lyubartsev AP (2012) Derivation and systematic validation of a refined all-atom force field for phosphatidylcholine lipids. J Phys Chem B 116:3164–3179
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97:10269–10280
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Pople JA et al (2010) GAUSSIAN 09 (revision B.01). Gaussian, Wallingford
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247260
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general AMBER force field. J Comput Chem 2:1157–1174
Hub JS, de Groot BL, van der Spoel D (2010) g_wham—a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Theory Comput 6:3713–3720
Small-Molecule Drug Discovery Suite 2015–1 (2015) QSite, version 6.6, Schrödinger, LLC, New York, NY
Murphy RB, Philipp DM, Friesner RA (2000) A mixed quantum mechanics/molecular mechanics (QM/MM) method for large-scale modeling of chemistry in protein environments. J Comp Chem 21:1442–1457
Hasegawa K, Noguchi T (2014) Molecular interactions of the quinone electron acceptors Q(A), Q(B), and Q(C) in photosystem II as studied by the fragment molecular orbital method. Photosynth Res 120:113–123
Kern J, Renger G (2007) Photosystem II: structure and mechanism of the water: plastoquinone oxidoreductase. Photosynth Res 94:183–202
Balabin IA, Onuchic JN (2000) Dynamically controlled protein tunneling paths in photosynthetic reaction centers. Science. 290:114–117
Nishioka H, Kimura A, Yamato T, Kawatsu T, Kakitani T (2005) Interference, fluctuation, and alternation of electron tunneling in protein media. 1. Two tunneling routes in photosynthetic reaction center alternate due to thermal fluctuation of protein conformation. J Phys Chem B 109(5):1978–1987
Ishikita H, Knapp EW (2005) Control of quinone redox potentials in photosystem II: electron transfer and photoprotection. J Am Chem Soc 127:14714–14720
Lambreva MD, Russo D, Polticelli F, Scognamiglio V, Antonacci A, Zobnina V, Campi G, Rea G (2014) Structure/function/dynamics of photosystem II plastoquinone binding sites. Curr Protein Pept Sci 15:285–295
Broser M, Glöckner C, Gabdulkhakov A, Guskov A, Buchta J, Kern J, Müh F, Dau H, Saenger W, Zouni A (2011) Structural basis of cyanobacterial photosystem II inhibition by the herbicide terbutryn. J Biol Chem 286:15964–15972
Graige MS, Paddock ML, Bruce JM, Feher G, Okamura MY (1996) Mechanism of proton-coupled electron transfer for quinone (Q B ) reduction in reaction centers of Rb. sphaeroides. J Am Chem Soc 118:9005–9016
Graige MS, Paddock ML, Feher G, Okamura MY (1999) Observation of the protonated semiquinone intermediate in isolated reaction centers from Rhodobacter sphaeroides: implications for the mechanism of electron and proton transfer in proteins. Biochemistry 38:11465–11473
Graige MS, Feher G, Okamura MY (1998) Conformational gating of the electron transfer reaction QA- QB –> QAQB- in bacterial reaction centers of Rhodobacter sphaeroides determined by a driving force assay. Proc Natl Acad Sci U S A. 95:11679–11684
Stowell MH, McPhillips TM, Rees DC, Soltis SM, Abresch E, Feher G (1997) Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer. Science. 276:812–816
Baxter RH, Ponomarenko N, Srajer V, Pahl R, Moffat K, Norris JR (2004) Time-resolved crystallographic studies of light-induced structural changes in the photosynthetic reaction center. Proc Natl Acad Sci U S A. 101:5982–5987
Reifarth F, Renger G (1998) Indirect evidence for structural changes coupled with QB- formation in photosystem II. FEBS Lett. 428:123–126
Alexov EG, Gunner MR (1999) Calculated protein and proton motions coupled to electron transfer: electron transfer from QA- to QB in bacterial photosynthetic reaction centers. Biochemistry 38:8253–8270
Saito K, Rutherford AW, Ishikita H (2013) Mechanism of proton-coupled quinone reduction in photosystem II. Proc Natl Acad Sci U S A. 110:954–959
Sundby C et al (1993) Effects on photosystem II function, photoinhibition, and plant performance of the spontaneous mutation of Serine-264 in the photosystem II reaction center D1 protein in triazine-resistant Brassica napus L. Plant Physiol. 103:105–113
Chernyshev A, Cukierman S (2002) Thermodynamic view of activation energies of proton transfer in various gramicidin A channels. Biophys J 82:182–192
Klauss A, Haumann M, Dau H (2012) Alternating electron and proton transfer steps in photosynthetic water oxidation. PNAS 109:16035–16040
Cox N, Jin L, Jaszewski A, Smith PJ, Krausz E, Rutherford AW, Pace R (2009) The semiquinone–iron complex of photosystem II: structural insights from ESR and theoretical simulation; evidence that the native ligand to the non-heme iron is carbonate. Biophys J 97:2024–2033
Ishikita H, Saito K (2013) Proton transfer reactions and hydrogen-bond networks in protein environments. J R Soc Interface 11:20130518
Shevela D, Eaton-Rye JJ, Shen JR, Govindjee (2012) Photosystem II and the unique role of bicarbonate: a historical perspective. Biochim Biophys Acta 1817:1134–1151
Xiong J, Subramaniam S, Govindjee (1996) Modeling of the D1/D2 proteins and cofactors of the photosystem II reaction center: implications for herbicide and bicarbonate binding. Protein Sci. 5:2054–2073
Funding
This work was supported by the Czech Science Foundation (project no GA15-12816S). Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the program “Projects of Large Research, Development, and Innovations Infrastructures” (CESNET LM2015042) is greatly appreciated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 2489 kb)
Rights and permissions
About this article
Cite this article
Kulik, N., Kutý, M. & Řeha, D. The study of conformational changes in photosystem II during a charge separation. J Mol Model 26, 75 (2020). https://doi.org/10.1007/s00894-020-4332-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-4332-9