Abstract
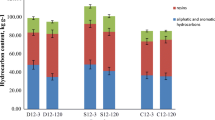
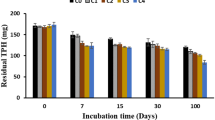
Crude oil extracted from oilfield reservoirs brings together hypersaline produced water. Failure in pipelines transporting this mixture causes contamination of the soil with oil and hypersaline water. Soil salinization is harmful to biological populations, impairing the biodegradation of contaminants. We simulated the contamination of a soil from an oilfield with produced water containing different concentrations of NaCl and crude oil, in order to evaluate the effect of salinity and hydrocarbon concentration on prokaryote community structure and biodegradation activity. Microcosms were incubated in CO2-measuring respirometer. After the incubation, residual aliphatic hydrocarbons were quantified and were performed 16S rRNA gene sequencing. An increase in CO2 emission and hydrocarbon biodegradation was observed with increasing oil concentration up to 100 g kg−1. Alpha diversity decreased in oil-contaminated soils with an increase in the relative abundance of Actinobacteria and reduction of Bacteroidetes with increasing oil concentration. In the NaCl-contaminated soils, alpha diversity, CO2 emission, and hydrocarbon biodegradation decreased with increasing NaCl concentration. There was an increase in the relative abundance of Firmicutes and Proteobacteria and a reduction of Actinobacteria with increasing salt concentration. Our results highlight the need to adopt specific bioremediation strategies in soils impacted by mixtures of crude oil and hypersaline produced water.








Similar content being viewed by others
References
Atoufi DH, Lampert DJ (2020) Impacts of oil and gas production on contaminant levels in sediments. Curr Pollut Rep 6:43–53. https://doi.org/10.1007/s40726-020-00137-5
Abdol Hamid HR, Kassim WM, El Hishir A, El-Jawashi SA (2008) Risk assessment and remediation suggestion of impacted soil by produced water associated with oil production. Environ Monit Assess 145(1-3):95–102. https://doi.org/10.1007/s10661-007-0018-3
Stewart, M., Arnold, K. (2011). Produced water treatment: field manual. part 1 - produced water treating systems, p. 1-134.
Hallsworth JE, Yakimov MM, Golyshin PN, Gillion JL, D’Auria G, de Lima Alves F, La Cono V, Genovese M, McKew BA, Hayes SL, Harris G, Giuliano L, Timmis KN, McGenity TJ (2007) Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ Microbiol 9(3):801–813. https://doi.org/10.1111/j.1462-2920.2006.01212.x
Stevenson A, Cray JA, Williams JP, Santos R, Sahay R, Neuenkirchen N, McClure CD, Grant IR, Houghton JDR, Quinn JP, Timson DJ, Patil SV, Singhal RS, Anton J, Dijksterhuis J, Hocking AD, Lievens B, Rangel DEN, Voytek MA, Gunde-Cimerman N, Oren A, Timmis KN, McGenity TJ, Hallsworth JE (2015) Is there a common water-activity limit for the three domains of life? ISME J 9:1333–1351. https://doi.org/10.1038/ismej.2014.219
Means JC (1995) Influence of salinity upon sediment-water partitioning of aromatic hydrocarbons. Mar Chem 51:3–16
Turner A, Rawling MC (2001) The influence of salting out on the sorption of neutral organic compounds in estuaries. Water Res 35(18):4379–4389. https://doi.org/10.1016/s0043-1354(01)00163-4
Mackay D, Shiu WY, Ma KC, Lee SC (2006) Handbook of physical-chemical and environmental fate for organic chemicals, 2nd edn. Taylor & Francis, Boca Raton
Pirnik MP, Atlas RM, Bartha R (1974) Hydrocarbon metabolism by Brevibacterium erythrogenes: normal and branched alkanes. J Bacteriol 119(3):868–878
Seklemova E, Pavlova A, Kovacheva K (2001) Biostimulation-based bioremediation of diesel fuel: field demonstration. Biodegradation 12(5):311–316. https://doi.org/10.1023/a:1014356223118
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Supplement1):4516–4522. https://doi.org/10.1073/pnas.1000080107
Pylro VS, Roesch L, Ortega JM, do Amaral, A. M. (2014) Brazilian microbiome project: revealing the unexplored microbial diversity—challenges and prospects. Microb Ecol 67:237–241. https://doi.org/10.1007/s00248-013-0302-4
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu F, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072. https://doi.org/10.1128/AEM.03006-05
Seedorf H, Kittelmann S, Henderson G, Janssen PH (2014) RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ 2:e494. https://doi.org/10.7717/peerj.494
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40(34):237–264
Chao A (1984) Nonparametric estimation of the numbers of classes in a population. Scand J Stat 11:265270
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:623–656
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71(12):8228–8235. https://doi.org/10.1128/AEM.71.12.8228-8235.2005
Franco I, Contin M, Bragato G, De Nobili M (2004) Microbiological resilience of soils contaminated with crude oil. Geoderma 121(1):17–30. https://doi.org/10.1016/j.geoderma.2003.10.002
Cury, J.D.E.C. (2002). Atividade microbiana e diversidades metabólica e genética em solo de mangue contaminado com petróleo. Master’s Dissertation. Universidade de São Paulo. São Paulo, Brazil.
Hernández-López EL, Ayala M, Vazquez-Duhalt R (2015) Microbial and enzymatic biotransformations of asphaltenes. Pet Sci Technol 33:1019–1027. https://doi.org/10.1080/10916466.2015.1014960
Atlas RM, Bartha R (1998) Microbial ecology: fundamentals and applications, 4th edn. Menlo Park, California 694 p
Fernández, P.S.H. (2016). Predação de protozoários sobre ultramicrocélulas bacterianas degradadoras de hidrocarbonetos e seu efeito sobre a biorremediação de agregados de solo contaminados com petróleo. Master’s Dissertation, Universidade Federal de Viçosa, Brasil.
El-Tarabily KA (2002) Total microbial activity composition of a mangrove sediment are reduced by oil pollution at a site in the Arabian Gulf. Can J Microbiol 48(2):176–182. https://doi.org/10.1139/w01-140
Labud V, Garcia C, Hernandez T (2007) Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere 66(10):1863–1871. https://doi.org/10.1016/j.chemosphere.2006.08.021
Camacho-Montealegre CM, Rodrigues EM, Tótola MR (2019) Microbial diversity and bioremediation of rhizospheric soils from Trindade Island - Brazil. J Environ Manag 236:358–364. https://doi.org/10.1016/j.jenvman.2019.02.013
Yang S, Wen X, Zhao L, Shi Y, Jin H (2014) Crude oil treatment leads to shift of bacterial communities in soils from the deep active layer and upper permafrost along the China-Russia Crude Oil Pipeline route. PLoS ONE 9(5):e96552. https://doi.org/10.1371/journal.pone.0096552
Atlas RM (1981) Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev 45:180–209
Rodrigues EM, Kalks KHM, Tótola MR (2015) Prospect, isolation, and characterization of microorganisms for potential use in cases of oil bioremediation along the coast of Trindade Island – Brazil. J Environ Manag 156:15–22. https://doi.org/10.1016/j.jenvman.2015.03.016
Kim JS, Crowley DE (2007) Microbial diversity in natural asphalts of the Rancho La Brea Tar Pits. Appl Environ Microbiol 73(14):45794591–45794591. https://doi.org/10.1128/AEM.01372-06
Chaturvedi S, Khurana SMP (2019) Importance of Actinobacteria for bioremediation. In: Khurana S, Gaur R (eds) Plant Biotechnology: Progress in Genomic Era. Springer, Singapore
Lang S, Philp JC (1998) Surface active lipids in rhodococci. Antonie Van Leuwenhoek 74:59–70
Balachandran C, Duraipandiyan V, Balakrishna K, Ignacimuthu S (2012) Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresour Technol 112:8390. https://doi.org/10.1016/j.biortech.2012.02.059
Morais D, Pylro V, Clark IM, Hirsch PR, Tótola MR (2016) Responses of microbial community from tropical pristine coastal soil to crude oil contamination. PeerJ 4:e1733. https://doi.org/10.7717/peerj.1733
McGowan L, Herbert R, Muyzer G (2004) A comparative study of hydrocarbon degradation by Marinobacter sp., Rhodococcus sp. and Corynebacterium sp. isolated from different mat systems. Ophelia 58(3):271–281. https://doi.org/10.1080/00785236.2004.10410235
Kurniati TH, Rusmana I, Suryani A, Mubarik NR (2016) Degradation of polycyclic aromatic hydrocarbon Pyrene by biosurfactant-producing bacteria Gordonia cholesterolivorans AMP 10. J Biol Biol Educ 8(3):336–343. https://doi.org/10.15294/biosaintifika.v8i3.6448
Rodrigues EM, Teixeira AVNC, Cesar DE, Tótola MR (2020) Strategy to improve crude oil biodegradation in oligotrophic aquatic environments: W/O/W fertilized emulsions and hydrocarbonoclastic bacteria. Braz J Microbiol 51:1159–1168. https://doi.org/10.1007/s42770-020-00244-x
Qiao N, Shao Z (2010) Isolation and characterization of a novel biosurfactant produced by hydrocarbon-degrading bacterium Alcanivorax dieselolei B-5. J Appl Microbiol 108(4):1207–1216. https://doi.org/10.1111/j.1365-2672.2009.04513.x
Da Silva FSP, Pylro VS, Fernandes PL, Barcelos GS, Kalks KHM, Schaefer CEGR, Tótola MR (2015) Unexplored Brazilian oceanic island host high salt tolerant biosurfactant-producing bacterial strains. Extremophiles 19(3):561–572. https://doi.org/10.1007/s00792-015-0740-7
Fernandes PL, Rodrigues EM, Paiva FR, Ayupe BAL, McInerney MJ, Tótola MR (2016) Biosurfactant, solvents and polymer production by Bacillus subtilis RI4914 and their application for enhanced oil recovery. Fuel 180:551–557. https://doi.org/10.1016/j.fuel.2016.04.080
Minai-Tehrani D, Herfatmanesh A, Azari-Dehkordi F, Minuoi S (2006) Effect of salinity on biodegradation of aliphatic fractions of crude oil in soil. Pak J Biol Sci 9(8):1531–1535 https://doi.org/10.3923/pjbs.2006.1531.1535
McGenity TJ (2010) Halophilic hydrocarbon degraders. In: Timmis KN (ed) Handbook of Hydrocarbon and Lipid Microbiology. Springer-Verlag, Berlin, pp 1939–1951
Ulrich AC, Guigard SE, Foght JM, Semple KM, Pooley K, Armstrong JE, Biggar KW (2009) Effect of salt on aerobic biodegradation of petroleum hydrocarbons in contaminated groundwater. Biodegradation 20(1):27–38. https://doi.org/10.1007/s10532-008-9196-0
Qin X, Tang JC, Li DS, Zhang QM (2012) Effect of salinity on the bioremediation of petroleum hydrocarbons in a saline-alkaline soil. Lett Appl Microbiol 55:210–217. https://doi.org/10.1111/j.1472-765X.2012.03280.x
Oren A (2002) Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol 28(1):56–63. https://doi.org/10.1038/sj/jim/7000176
Yakimov MM, Golyshin PN, Lang S, Moore ER, Abraham WR, Lünsdorf H, Timmis KN (1998) Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int J Syst Evol Microbiol 48:339–348. https://doi.org/10.1099/00207713-48-2-339
Syutsubo K, Kishira H, Harayama S (2001) Development of specific oligonucleotide probes for the identification and in situ detection of hydrocarbon-degrading Alcanivorax strains. Environ Microbiol 3:371–379. https://doi.org/10.1046/j.1462-2920.2001.00204.x
Hara A, Syutsubo K, Harayama S (2003) Alcanivorax which prevails in oil contaminated sea water exhibits broad substrate specificity for alkane degradation. Environ Microbiol 5:746–753. https://doi.org/10.1046/j.1468-2920.2003.00468.x
Scoma A, Boon N (2016) Osmotic stress confers enhanced cell integrity to hydrostatic pressure but impairs growth in Alcanivorax borkumensis SK2. Front Microbiol 7:729. https://doi.org/10.3389/fmicb.2016.00729
Dastgheib SMM, Amoozegar MA, Khajeh K, Ventosa A (2011) A halotolerant Alcanivorax sp. strain with potential application in saline soil remediation. Appl Microbiol Biotechnol 90:305–312. https://doi.org/10.1007/s00253-010-3049-6
Gu J, Cai H, Yu SL, Qu R, Yin B, Guo YF, Zhao JY, Wu XL (2007) Marinobacter gudaonensis sp. nov., isolated from an oil-polluted saline soil in a Chinese oilfield. Int J Syst Evol Microbiol 57(Pt 2):250–254. https://doi.org/10.1099/ijs.0.64522-0
Gomes MB, Gonzales-Limache EE, Sousa STP, Dellagnezze BM, Sartoratto A, Silva LCF, Gieg LM, Valoni E, Souza RS, Torres APR, Sousa MP, De Paula SO, Silva CC, Oliveira VM (2016) Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int Biodeterior Biodegrad 126:231–242. https://doi.org/10.1016/j.ibiod.2016.08.014
Al-Mailem DM, Al-Deieg M, Eliyas M, Radwan SS (2017) Biostimulation of indigenous microorganisms for bioremediation of oily hypersaline microcosms from the Arabian Gulf Kuwaiti coasts. J Environ Manag 193:576–583 https://doi.org/10.1016/j.jenvman.2017.02.054
Kuhlmann AU, Bremer E (2002) Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl Environ Microbiol 68:772–783. https://doi.org/10.1128/AEM.68.2.772-783.2002
Höper D, Bernhardt J, Hecker M (2006) Salt stress adaptation of Bacillus subtilis: a physiological proteomics approach. Proteomics 6:1550–1562. https://doi.org/10.1002/pmic.200500197
Ghani M, Ansari A, Aman A, Zohra RR, Siddiqui Nn, Qader SA (2013) Isolation and characterization of different strains of Bacillus licheniformis for the production of commercially significant enzymes. Pak J Pharm Sci 26(4):691–697
Hui Y, Huang G, An C, Wei J (2011) Combined effects of DOM extracted from site soil/compost and biosurfactant on the sorption and desorption of PAHs in a soil-water system. J Hazard Mater 190(1-3):883–890. https://doi.org/10.1016/j.jhazmat.2011.04.026
Bezza FA, Chirwa EMN (2016) Biosurfactant-enhanced bioremediation of aged polycyclic aromatic hydrocarbons (PAHs) in creosote contaminated soil. Chemosphere 144:635–644. https://doi.org/10.1016/j.chemosphere.2015.08.027
Koenigsberg SS, Sandefur CA (1999) The use of oxygen release compound for the accelerated bioremediation of aerobically degradable contaminants: the advent of time-release electron acceptors. Remediat J 10:3–29
Zhao B, Wang H, Mao X, Li R (2009) Biodegradation of phenanthrene by a halophilic bacterial consortium under aerobic conditions. Curr Microbiol 58:205–210. https://doi.org/10.1007/s00284-008-9309-3
Wang, X., Zheng, J., Han, Z., Chen, H., 2019. Bioremediation of crude oil-contaminatedsoil by hydrocarbon-degrading microorganisms immobilized on humic acid-modifiedbiofuel ash. Journal of Chemical Technology, Biot.10.1002/jctb.5969.
Martins LF, Peixoto RS (2012) Biodegradation of petroleum hydrocarbons in hypersaline environments. Braz J Microbiol 43:865–872. https://doi.org/10.1590/S1517-83822012000300003
Acknowledgements
We are thankful to the Brazilian Navy, FAPEMIG, and CAPES.
Funding
This work was supported by National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Acacio Aparecido Navarrete
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 253 kb)
Rights and permissions
About this article
Cite this article
Camacho-Montealegre, C.M., Rodrigues, E.M., Morais, D.K. et al. Prokaryotic community diversity during bioremediation of crude oil contaminated oilfield soil: effects of hydrocarbon concentration and salinity. Braz J Microbiol 52, 787–800 (2021). https://doi.org/10.1007/s42770-021-00476-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00476-5