Abstract
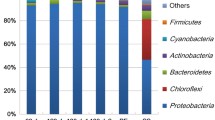
A robust and widely applicable method for sampling of aquatic microbial biofilm and further sample processing is presented. The method is based on next-generation sequencing of V4–V5 variable regions of 16S rRNA gene and further statistical analysis of sequencing data, which could be useful not only to investigate taxonomic composition of biofilm bacterial consortia but also to assess aquatic ecosystem health. Five artificial materials commonly used for biofilm growth (glass, stainless steel, aluminum, polypropylene, polyethylene) were tested to determine the one giving most robust and reproducible results. The effect of used sampler material on total microbial composition was not statistically significant; however, the non-plastic materials (glass, metal) gave more stable outputs without irregularities among sample parallels. The bias of the method is assessed with respect to the employment of a non-quantitative step (PCR amplification) to obtain quantitative results (relative abundance of identified taxa). This aspect is often overlooked in ecological and medical studies. We document that sequencing of a mixture of three merged primary PCR reactions for each sample and further evaluation of median values from three technical replicates for each sample enables to overcome this bias and gives robust and repeatable results well distinguishing among sampling localities and seasons.






Similar content being viewed by others
Abbreviations
- NGS:
-
Next-generation sequencing
- NMDS:
-
Non-metric multidimensional scaling
- DGGE:
-
Denaturing gradient gel electrophoresis
- T-RFLP:
-
Terminal restriction fragment length polymorphism
- FISH:
-
Fluorescent in situ hybridization
- POCIS:
-
Polar organic chemical integrative sampler
- G:
-
Basic soda lime silicate glass
- S:
-
Stainless steel
- A:
-
Aluminum
- PP:
-
Polypropylene
- PE:
-
High-density polyethylene
- STP:
-
Sewage treatment plant
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ammar Y, Swailes D, Bridgens B, Chen JJ (2015) Influence of surface roughness on the initial formation of biofilm. Surf Coat Technol 284:410–416
Araya R, Tani K, Takagi T, Yamaguchi N, Nasu M (2003) Bacterial activity and community composition in stream water and biofilm from an urban river determined by fluorescent in situ hybridization and DGGE analysis. FEMS Microbiol Ecol 43:111–119
Aronesty E (2013) Comparison of sequencing utility programs. Open Bioinforma J 7:1–8
Balcazar JL, Subirats J, Borrego CM (2015) The role of biofilms as environmental reservoirs of antibiotic resistance. Front Microbiol 6:1–9
Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T, Zifcakova L, Snajdr J, Ridl J, Vlcek C, Voriskova J (2012) Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6:248–258
Battin TJ, Kaplan LA, Findlay S, Hopkinson CS, Marti E, Packman AI, Newbold JD, Sabater F (2008) Biophysical controls on organic carbon fluxes in fluvial networks. Nat Geosci 1:95–100
Battin TJ, Besemer K, Bengtsson MM, Romani AM, Packmann AI (2016) The ecology and biogeochemistry of stream biofilms. Nat Rev Microbiol 14:251–263
Besemer K (2015) Biodiversity, community structure and function of biofilms in stream ecosystems. Res Microbiol 166:774–781
Besemer K, Peter H, Logue JB, Langenheder S, Lindstrom ES, Tranvik LJ, Battin TJ (2012) Unraveling assembly of stream biofilm communities. ISME J 6:1459–1468
Drury B, Rosi-Marshall E, Kelly JJ (2013) Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl Environ Microbiol 79:1897–1905
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Grabicova K, Grabic R, Blaha M, Kumar V, Cerveny D, Fedorova G, Randak T (2015) Presence of pharmaceuticals in benthic fauna living in a small stream affected by effluent from a municipal sewage treatment plant. Water Res 72:145–153
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis., Palaeontologia electronica. http://palaeo-electronica.org. Coquina Press, 4:1–9
Hellal J, Michel C, Barsotti V, Laperche V, Garrido F, Joulian C (2016) Representative sampling of natural biofilms: influence of substratum type on the bacterial and fungal communities structure. Springerplus 5:1–9
Hoellein T, Rojas M, Pink A, Gasior J, Kelly J (2014) Anthropogenic litter in urban freshwater ecosystems: distribution and microbial interactions. PLoS One 9:1–13
Lau KEM, Washington VJ, Fan V, Neale MW, Lear G, Curran J, Lewis GD (2015) A novel bacterial community index to assess stream ecological health. Freshw Biol 60:1988–2002
Loza V, Perona E, Mateo P (2013) Molecular fingerprinting of Cyanobacteria from river biofilms as a water quality monitoring tool. Appl Environ Microbiol 79:1459–1472
Muyzer G, Teske A, Wirsen CO, Jannasch HW (1995) Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164:165–172
Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, Fire AZ (2007) A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35:1–9
Pilloni G, Granitsiotis MS, Engel M, Lueders T (2012) Testing the limits of 454 Pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS One 7:1–7
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596
R Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Reuben S, Chua CLN, Fam KD, Thian ZYA, Kang MK, Swarup S (2012) Bacterial diversity on different surfaces in urban freshwater. Water Sci Technol 65:1869–1874
Romani AM, Guasch H, Munoz I, Ruana J, Vilalta E, Schwartz T, Emtiazi F, Sabater S (2004) Biofilm structure and function and possible implications for riverine DOC dynamics. Microb Ecol 47:316–328
Samarajeewa AD, Hammad A, Masson L, Khan IUH, Scroggins R, Beaudette LA (2015) Comparative assessment of next-generation sequencing, denaturing gradient gel electrophoresis, clonal restriction fragment length polymorphism and cloning-sequencing as methods for characterizing commercial microbial consortia. J Microbiol Methods 108:103–111
Turner S, Pryer KM, Miao VPW, Palmer JD (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small submit rRNA sequence analysis. J Eukaryot Microbiol 46:327–338
Vetrovsky T, Baldrian P (2013a) Analysis of soil fungal communities by amplicon pyrosequencing: current approaches to data analysis and the introduction of the pipeline SEED. Biol Fertil Soils 49:1027–1037
Vetrovsky T, Baldrian P (2013b) The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:1–10
Wakelin SA, Colloff MJ, Kookana RS (2008) Effect of wastewater treatment plant effluent on microbial function and community structure in the sediment of a freshwater stream with variable seasonal flow. Appl Environ Microbiol 74:2659–2668
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO (2014) The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:643–648
Zeglin LH (2015) Stream microbial diversity in response to environmental changes: review and synthesis of existing research. Front Microbiol 6:1–15
Zwart G, Crump BC, Agterveld M, Hagen F, Han SK (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155
Funding
This work was supported by the Czech Science Foundation (No. 15-04258S) and Ministry of Education, Youth and Sports of CR within the LQ1604 National Sustainability Program II (Project BIOCEV-FAR) and by the project “BIOCEV” (CZ.1.05/1.1.00/02.0109) from the European Regional Development Fund in the Czech Republic.
The financial support was also given to R. Grabic and V. Zlabek by the Ministry of Education, Youth and Sports of the Czech Republic, projects CENAKVA (No. CZ.1.05/2.1.00/01.0024) and CENAKVA II (No. LO1205 under the NPU I program).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Online Resource 1
The list of used composite primers sequences. (XLSX 11 kb)
Online Resource 2
The list of identified taxons (cutoff 0.5%) in experimental and natural samples. (XLSX 59 kb)
Online Resource 3
The number of identified taxons (cutoffs 0.5, 1, 2%) in experimental and natural samples. (XLSX 12 kb)
Online Resource 4
The Bray-Curtis distance matrix calculated for individual experimental and natural samples (cutoff 1%). (XLSX 29 kb)
Online Resource 5
NMDS plot of Bray-Curtis distance matrix for individual experimental samples and medians of natural samples; taxon relative abundance cutoff 0.5% in at least one sample (stress value = 0.149). The values for samples of the experimental set are marked as follows: circle = G; square = A; diamond = PE, triangle = PP; inv. triangle = S. Filled shapes stand for non-rinsed samples, blank ones for rinsed samples. The red star represents the absolute median of all values of the experimental sample set. Natural samples from Zivny stream are marked as follows: dash = upstream STP, cross = downstream STP, gray color = spring, blue = autumn. (PDF 88 kb)
Online Resource 6
NMDS plot of Bray-Curtis distance matrix for individual experimental samples and medians of natural samples; taxon relative abundance cutoff 2% in at least one sample (stress value = 0.137). The values for samples of the experimental set are marked as follows: circle = G; square = A; diamond = PE, triangle = PP; inv. triangle = S. Filled shapes stand for non-rinsed samples, blank ones for rinsed samples. The red star represents the absolute median of all values of the experimental sample set. Natural samples from Zivny stream are marked as follows: Dash = upstream STP, cross = downstream STP, gray color = spring, blue = autumn. (PDF 90 kb)
Online Resource 7
The Bray-Curtis distance matrix calculated for medians from triplicates of experimental and natural samples (cutoff 2%). (XLSX 12 kb)
Online Resource 8
The Bray-Curtis distance matrix calculated for medians from triplicates of experimental and natural samples (cutoff 1%). (XLSX 13 kb)
Online Resource 9
The Bray-Curtis distance matrix calculated for medians from triplicates of experimental and natural samples (cutoff 0.5%). (XLSX 12 kb)
Online Resource 10
The Bray-Curtis distance matrix of comparative samples of medians calculated for triplicates of testing set samples to the absolute median of testing set samples (median calculated for all 30 testing samples) for all three cut-off levels 0.5, 1, and 2% (XLSX 9 kb)
Online Resource 11
The source data for Venn diagrams (Fig. 6). (XLSX 339 kb)
Rights and permissions
About this article
Cite this article
Bakal, T., Janata, J., Sabova, L. et al. Suitability and setup of next-generation sequencing-based method for taxonomic characterization of aquatic microbial biofilm. Folia Microbiol 64, 9–17 (2019). https://doi.org/10.1007/s12223-018-0624-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-018-0624-1