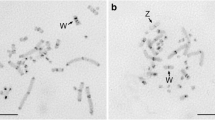
Abstract
The W chromosome of most lepidopteran species represents the largest heterochromatin entity in the female genome. Although satellite DNA is a typical component of constitutive heterochromatin, there are only a few known satellite DNAs (satDNAs) located on the W chromosome in moths and butterflies. In this study, we isolated and characterized new satDNA (PiSAT1) from microdissected W chromosomes of the Indian meal moth, Plodia interpunctella. Even though the PiSAT1 is mainly localized near the female-specific segment of the W chromosome, short arrays of this satDNA also occur on autosomes and/or the Z chromosome. Probably due to the predominant location in the non-recombining part of the genome, PiSAT1 exhibits a relatively large nucleotide variability in its monomers. However, at least a part of all predicted functional motifs is located in conserved regions. Moreover, we detected polyadenylated transcripts of PiSAT1 in all developmental stages and in both sexes (female and male larvae, pupae and adults). Our results suggest a potential structural and functional role of PiSAT1 in the P. interpunctella genome, which is consistent with accumulating evidence for the important role of satDNAs in eukaryotic genomes.






Similar content being viewed by others
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- cDNA:
-
Complementary DNA
- CGH:
-
Comparative genomic hybridization
- DABCO:
-
1,4-Diazabicyclo(2.2.2)-octane
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- gDNA:
-
Genomic DNA
- LTR:
-
Long terminal repeats
- mtDNA:
-
Mitochondrial DNA
- ncRNA:
-
Non-coding RNA
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- RNAi:
-
RNA interference
- satDNA:
-
Satellite DNA
- SDS:
-
Sodium dodecyl sulphate
- SSC:
-
Saline-sodium citrate buffer
References
Abe H, Mita K, Yasukochi Y, Oshiki T, Shimada T (2005) Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet Genome Res 110:144–151. doi:10.1159/000084946
Biscotti MA, Barucca M, Capriglione T, Odierna G, Olmo E, Canapa A (2008) Molecular and cytogenetic characterization of repetitive DNA in the Antarctic polyplacophoran Nuttallochiton mirandus. Chromosom Res 16:907–916. doi:10.1007/s10577-008-1248-0
Biscotti MA, Canapa A, Forconi M, Olmo E, Barucca M (2015a) Transcription of tandemly repetitive DNA: functional roles. Chromosom Res 23:463–477. doi:10.1007/s10577-015-9494-4
Biscotti MA, Olmo E, Heslop-Harrison JSP (2015b) Repetitive DNA in eukaryotic genomes. Chromosom Res 23:415–420. doi:10.1007/s10577-015-9499-z
Casanova M, Pasternak M, ElMarjou F, LeBaccon P, Probst AV, Almouzni G (2013) Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep 4:1156–1167. doi:10.1016/j.celrep.2013.08.015
Charlesworth B, Charlesworth D (2000) The degeneration of Y chromosomes. Philos Trans R Soc B Biol Sci 355:1563–1572. doi:10.1098/rstb.2000.0717
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220. doi:10.1038/371215a0
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb) 95:118–128. doi:10.1038/sj.hdy.6800697
Ferreira D, Meles S, Escudeiro A, Mendes-da-Silva A, Adega F, Chaves R (2015) Satellite non-coding RNAs: the emerging players in cells, cellular pathways and cancer. Chromosom Res 23:479–493. doi:10.1007/s10577-015-9482-8
Frydrychová R, Marec F (2002) Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera). Genetica 115:179–187. doi:10.1023/A:1020175912128
Fuková I, Nguyen P, Marec F (2005) Codling moth cytogenetics: karyotype, chromosomal location of rDNA, and molecular differentiation of sex chromosomes. Genome 48:1083–1092. doi:10.1139/g05-063
Fuková I, Traut W, Vítková M, Nguyen P, Kubíčková S, Marec F et al (2007) Probing the W chromosome of the codling moth, Cydia pomonella, with sequences from microdissected sex chromatin. Chromosoma 116:135–145. doi:10.1007/s00412-006-0086-0
Gabrielian A, Vlahovicek K, Pongor S (1997) Distribution of sequence-dependent curvature in genomic DNA sequences. FEBS Lett 406:69–74. doi:10.1016/S0014-5793(97)00236-6
Glaser RW (1917) The growth of insect blood cells in vitro. Psyche A J Entomol 24:1–7. doi:10.1155/1917/67649
King LM, Cummings MP (1997) Satellite DNA repeat sequence variation is low in three species of burying beetles in the genus Nicrophorus (Coleoptera: Silphidae). Mol Biol Evol 14:1088–1095
Kubickova S, Cernohorska H, Musilova P, Rubes J (2002) The use of laser microdissection for the preparation of chromosome-specific painting probes in farm animals. Chromosom Res 10:571–577. doi:10.1023/A:1020914702767
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi:10.1093/bioinformatics/btp187
Lobov IB, Tsutsui K, Mitchell AR, Podgornaya OI (2001) Specificity of SAF-A and lamin B binding in vitro correlates with the satellite DNA bending state. J Cell Biochem 83:218–229. doi:10.1002/jcb.1220
Lockwood APM (1961) “Ringer”, solutions and some notes on the physiological basis of their ionic composition. Comp Biochem Physiol 2:241–289. doi:10.1016/0010-406X(61)90113-X
Lu Y-J, Kochert GD, Isenhour DJ, Adang MJ (1994) Molecular characterization of a strain-specific repeated DNA sequence in the fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Insect Mol Biol 3:123–130. doi:10.1111/j.1365-2583.1994.tb00159.x
Mahendran B, Acharya C, Dash R, Ghosh SK, Kundu SC (2006) Repetitive DNA in tropical tasar silkworm Antheraea mylitta. Gene 370:51–57. doi:10.1016/j.gene.2005.11.010
Mandrioli M, Manicardi GC, Marec F (2003) Cytogenetic and molecular characterization of the MBSAT1 satellite DNA in holokinetic chromosomes of the cabbage moth, Mamestra brassicae (Lepidoptera). Chromosom Res 11:51–56. doi:10.1023/A:1022058032217
Mediouni J, Fuková I, Frydrychová R, Dhouibi MH, Marec F (2004) Karyotype, sex chromatin and sex chromosome differentiation in the carob moth, Ectomyelois ceratoniae (Lepidoptera: Pyralidae). Caryologia 57:184–194. doi:10.1080/00087114.2004.10589391
Mravinac B, Plohl M, Ugarković D (2004) Conserved patterns in the evolution of Tribolium satellite DNAs. Gene 332:169–177. doi:10.1016/j.gene.2004.02.055
Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H (2008) Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol 38:1046–1057. doi:10.1016/j.ibmb.2008.05.012
Palomeque T, Lorite P (2008) Satellite DNA in insects: a review. Heredity (Edinb) 100:564–573. doi:10.1038/hdy.2008.24
Pavlek M, Gelfand Y, Plohl M, Meštrović N (2015) Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Res 22:387–401. doi:10.1093/dnares/dsv021
Petraccioli A, Odierna G, Capriglione T, Barucca M, Forconi M, Olmo E et al (2015) A novel satellite DNA isolated in Pecten jacobaeus shows high sequence similarity among molluscs. Mol Gen Genomics 290:1717–1725. doi:10.1007/s00438-015-1036-4
Pezer Ž, Brajković J, Feliciello I, Ugarković Đ (2011) Transcription of satellite DNAs in insects. In: Ugarković Ð (ed) Long non-coding RNAs, progress in molecular and subcellular biology, vol 51. Springer, Berlin, pp 161–178
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56. doi:10.1016/S0097-8485(01)00099-7
Rozas J, Rozas R (1995) DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Bioinformatics 11:621–625. doi:10.1093/bioinformatics/11.6.621
Sahara K, Marec F, Traut W (1999) TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosom Res 7:449–460. doi:10.1023/A:1009297729547
Sahara K, Marec F, Eickhoff U, Traut W (2003) Moth sex chromatin probed by comparative genomic hybridization (CGH). Genome 46:339–342. doi:10.1139/g03-003
Sahara K, Yoshido A, Traut W (2012) Sex chromosome evolution in moths and butterflies. Chromosom Res 20:83–94. doi:10.1007/s10577-011-9262-z
Satović E, Vojvoda Zeljko T, Luchetti A, Mantovani B, Plohl M (2016) Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements. BMC Genomics 17:997. doi:10.1186/s12864-016-3347-1
Šíchová J, Nguyen P, Dalíková M, Marec F (2013) Chromosomal evolution in tortricid moths: conserved karyotypes with diverged features. PLoS One 8:e64520. doi:10.1371/journal.pone.0064520
Simpson RT (1991) Nucleosome positioning: occurrence, mechanisms, and functional consequences. In: Cohn WE, Moldave K (eds) Progress in nucleic acid research and molecular biology. Academic Press, New York, pp 143–184
The International Silkworm Genome Consortium (2008) The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol 38:1036–1045. doi:10.1016/j.ibmb.2008.11.004
Traut W, Sahara K, Otto TD, Marec F (1999) Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma 108:173–180. doi:10.1007/s004120050366
Traut W, Sahara K, Marec F (2007) Sex chromosomes and sex determination in Lepidoptera. Sex Dev 1:332–346. doi:10.1159/000111765
Traut W, Vogel H, Glöckner G, Hartmann E, Heckel DG (2013) High-throughput sequencing of a single chromosome: a moth W chromosome. Chromosom Res 21:491–505. doi:10.1007/s10577-013-9376-6
Trick M, Dover GA (1984) Unexpectedly slow homogenisation within a repetitive DNA family shared between two subspecies of tsetse fly. J Mol Evol 20:322–329. doi:10.1007/BF02104738
Věchtová P, Dalíková M, Sýkorová M, Žurovcová M, Füssy Z, Zrzavá M (2016) CpSAT-1, a transcribed satellite sequence from the codling moth, Cydia pomonella. Genetica 144:385–395. doi:10.1007/s10709-016-9907-0
Viegas-Pequignot E, Dutrillaux B, Magdelenat H, Coppey-Moisan M (1989) Mapping of single-copy DNA sequences on human chromosomes by in situ hybridization with biotinylated probes: enhancement of detection sensitivity by intensified-fluorescence digital-imaging microscopy. Proc Natl Acad Sci U S A 86:582–586
Vítková M, Fuková I, Kubíčková S, Marec F (2007) Molecular divergence of the W chromosomes in pyralid moths (Lepidoptera). Chromosom Res 15:917–930. doi:10.1007/s10577-007-1173-7
Vlahoviček K, Kaján L, Pongor S (2003) DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Res 31:3686–3687. doi:10.1093/nar/gkg559
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. doi:10.1126/science.1074973
Acknowledgements
We thank Marie Korchová for the care of P. interpunctella cultures. Our thanks belong to Martina Žurovcová, Hanka Zikmundová, Sander Visser and M.Z. for providing P. interpunctella specimens to establish laboratory rearing. We are also thankful to Professor Steve Paterson for providing important information about the P. interpunctella genome assembly. This research was funded by grant 14-22765S of the Czech Science Foundation (GACR). Current support from GACR grants 17-17211S (M.D.) and 17-13713S (M.Z. and F.M.) is also gratefully acknowledged. S.K. acknowledges support from the Ministry of Education, Youth and Sports of the Czech Republic (CEITEC 2020 project LQ1601).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Irina Solovei
Electronic supplementary material
Online resource 1
Highly polyploid nuclei from female Malpighian tubules of Plodia interpunctella with heterochromatic body formed by multiple copies of W chromosome (indicated by arrows). Scale bars = 20 μm. (a) Part of polyploid nucleus from spreading preparation stained by Giemsa. (a) Part of Malpighian tubule from squash preparation stained by lactic acetic orcein. (PDF 351 kb)
Online resource 2
Alignment of PiSAT1 monomers against the consensus sequence. Sequences are listed in alphabetical order. PiSAT1a-d (GenBank accession number KY450690) are monomers from the original W chromosome-derived clone, PiSAT1 monomer 1-17 (GenBank accession numbers KY450691-KY450707) are additionally PCR isolated sequences, monomers obtained from Plodia interpunctella genome are named according to the respective scaffold. Red bars indicate places of alignment with indels longer than 2 bp. Grey bars refer to conserved regions of monomers based on sliding window analysis. *PiSAT1 monomers with deletions longer than 2 bp. +PiSAT1 monomers with insertions longer than 2 bp. (PDF 1090 kb)
Rights and permissions
About this article
Cite this article
Dalíková, M., Zrzavá, M., Kubíčková, S. et al. W-enriched satellite sequence in the Indian meal moth, Plodia interpunctella (Lepidoptera, Pyralidae). Chromosome Res 25, 241–252 (2017). https://doi.org/10.1007/s10577-017-9558-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-017-9558-8