1. Introduction
Research data management addresses the need for consistent data representation by applying the FAIR (Findable, Accessible, Interoperable, and Reusable) principles. In addition, this can be helped by ontologies, particularly for representing the data structure in the specific scientific domain [
1,
2]. Ontologies represent “a formal specification of a shared conceptualization” [
3]. By using a standardized and formalized description language, knowledge is expressed. Data are also enabled to be stored in a formalized, standardized description language. Here, terms used and relations between those terms can be specified, enabling for, among others, higher findability of data in databases. As different ontologies are created by different people, they are often not compatible, even within the same domain. Ontology creation and maintenance is a manual process, often performed by several experts. As each expert might work on different issues in their scientific work, they also may create conceptualizations of their respective knowledge, different from the conceptualizations provided by other domain experts. However, despite efforts to standardize knowledge conceptualization, there are often still multiple ontologies within the same domain [
4,
5].
Ontologies are applied in simple use cases as translation references in domain-specific vocabularies but also serve more complex purposes such as to serve as environments for property inference by logical reasoning. They define sets of representational primitives with which to model a domain of knowledge or discourse. The representational primitives mostly are classes, relationships, and their properties, e.g., definitions. Information elucidating their meaning is included as textual information in definition annotations of these primitives. In some, rather ideal, ontologies, logical consistency constraints with respect to the application are posed, as well [
6,
7]. Thus, a class can be defined in at least two different ways—through annotation with textual definitions, or as a product of its constraints and relations to other classes. Domain ontologies typically use domain-specific definitions of terms denoting their primitives.
As systems that rely on domain ontology interoperability become more extensive, it is often necessary to merge domain ontologies through manual reconciliation. This holds for enriching an ontology with information mined from domain-related texts. Approaches to automating knowledge conceptualization have limitations, as they require human user input to create semantics. Merging and extending ontologies is therefore a quite manual process that is both time-intensive and expensive.
Another problem domain experts face is ontology selection for a certain task. Different ontologies can focus on different levels of abstraction or place an emphasis on different sub-domains. Selection of the ontology, which best corresponds to a given document is an important part of reasoning about and increasing the FAIR-ness of such a text. Finding a suitable ontology for a given text database can speed up the process and help in classifying the information presented in the text. Moreover, the given text database is more easily connected to the data already linked within the ontology. An automated selection of ontologies and the respective classification of the text is thus desired, as this allows for the automated comparison of different text documents. In addition, the documents are compared in an understandable way and enable connection with the corresponding research data, which are also derived from other databases.
Ontology recommenders, e.g., the NCBO ontology recommender [
8], rate a text based on the presence of input text tokens. The similarity of tokens to preferred and alternate labels of ontology classes is calculated and is weighted by term frequency. At the same time, this work aims to use text embeddings instead in order to not only look for shared tokens but also to find concepts with synonymous meanings in text and ontology.
This article is dedicated to a particular challenge, sometimes encountered when enriching ontologies and their merging: deciding which of the some given ontologies is the most relevant to an input domain-related text fragment. The proposed method relies primarily on artificial neural networks (ANNs), specifically on using them in the Natural Language Processing (NLP) setting. To our best knowledge, the neural networks have never been used in combination with NLP for scoring the relevance of ontologies.
Since we propose several methods of assigning ontologies to input texts, it is needed to establish the difference between the proposed models. We do so through a comparison of the model performance on the ontology classification task, and conducting statistical tests on validation subsets to test the following five statistical hypotheses:
All six models perform the same on the validation splits.
The 10-NN models perform the same as their 1-NN variants.
The neural-network model performs the same as the k-NN model on BERT embeddings and on fastText embeddings.
For each of the considered models: The fine-tuned BERT with negative sampling has the same performance as other considered models.
For each remaining pair: Both models from the pair have the same performance.
The next section summarizes the related work on the usage of artificial neural networks (ANNs) on ontologies. In
Section 3, we recall the used methods of text preprocessing and embedding, as well as the summary of the used classifiers. The usage of the proposed methodology is described and evaluated in
Section 4. Finally, a discussion and an outlook for future work is given in
Section 5.
2. Related Work
In learning and extending ontologies, ANNs have been used mainly for identifying concepts, attributes, and their relations [
9,
10,
11]. Regarding relations, some ANN-based methods have been developed specifically for subsumption relations needed for taxonomy creation [
12,
13,
14,
15]. In the context of ontology integration, they were mainly used for ontology matching or ontology alignment [
16,
17,
18,
19]. The variety of the types of ANNs employed is quite large. It includes adaptive resonance theory (ART) networks [
20] and associative memories [
21], as well as multi-layer perceptron (MLP) [
22] and the modern deep convolutional neural network (CNN) [
16,
23], deep belief networks [
9], long-short term memory (LSTM) and bididectional long-short term memory (BiLSTM) [
24], and gated recurrent units (GRU) networks [
25,
26]. The dependency of ontologies on texts has led to the use of networks designed for learning texts and natural language representations. In particular, BERT [
27,
28] is a bidirectional encoder of representations from transformers.
Another important network is word2vec [
29], a rather traditional network used to embed the text into the Euclidean space. Originally designed for knowledge graphs [
30], RDF2Vec [
14,
26] was developed as a result of the close relationship between ontologies and knowledge graphs. With respect to vectorization networks such as word2vec and RDF2Vec, it is important to note that the OWL2Vec network was designed according to similar principles for embedding ontologies [
31]. Finally, the graph structure of ontologies has led to the use of graph neural networks (GNNs) [
18,
19].
The closest to our work is the way ANNs have recently been used in the context of OWL translation [
25,
32], in the linking and concatenation of predicates and restrictions [
21], and in extracting taxonomy from knowledge graphs [
14]. In the publication [
32], ontology learning is adapted as a transductive reasoning task that uses two recurrent neural networks to convert the natural language text into the description logic in web ontology language (OWL) specifications.
This approach was further developed in [
25] and resulted in a system that is based on a single recurrent GRU-type network. Employing an encoder-decoder architecture, it translates a subset of the natural language by the syntactic transformation into the description logic language ALLQ. It also generalizes across different syntactic structures and is able to handle unknown words by duplicating input words into the output as extralogical symbols and enriching the training set with new annotated examples.
A mapping between ontologies and a pair of mutually associative repositories is created in [
21], where one stores assertions and the other stores inference rules. In the recent work [
14] is described a methodology for the task of extracting a taxonomy from an embedded knowledge graph. The embedding can be obtained using, e.g., RDF2Vec.
Hierarchical agglomerative clustering is performed on this embedding, initially without using type information. Then, types are injected into the hierarchical cluster tree. Next, an axiom induction algorithm is applied to each cluster in the resulting tree to find new classes corresponding to those axioms that accurately describe the respective clusters.
3. Methodological Background
Details of the background of the methods employed in the reported research are described in this section. In the first part, we define the problem of matching texts to ontologies. In the second part, we describe the considered similarity measures. In the third part, we describe how to extract content from text documents, parse it into paragraphs and filter them to keep the minimal length and to be content relevant to the topic of the document. The fourth part describes the usage of a transformer for embedding input paragraphs into a vector space, where the classification will subsequently be performed. The final part describes the used classifiers.
3.1. Problem Definition of Matching Texts to Ontologies
Within the proposed framework, we define an ontology O as a directed attributed multi-graph, where vertices represent classes, edges represent relationships between them, and both vertices and edges can contain attributes. Given a set of specific ontologies and an input text , where is the space of all text documents. The target is to determine the ontology that best matches the content of T. To this end, we can employ a “hard” mapping or a scoring function which allows one to order ontologies by relevance.
The following two difficulties, which arise from the posed task, should be considered in the first place when choosing a solving method:
Given ontologies are the only source of supervision. No ground truth with pairs of source text and ontology labels are provided.
Ontologies may significantly differ in size. This can lead to very unbalanced datasets.
3.2. Matching Background
The above problem is closely related to the concept normalization and entity linking tasks, and the solutions include dictionary search [
33,
34], conditional random fields, and term frequency-inverse document frequency (tf-idf) vector similarity [
35], word embeddings, and syntactic similarity [
36].
Vector similarity approaches use either dense word embeddings or tf-idf vectors. The tf-idf vector is a document-specific vector, whose size is the size of the vocabulary under consideration. Each element in this vector is the count of occurrences of the term in a document multiplied by the logarithmized inverse of the proportion of documents in which the term occurs. These vectors are readily interpretable (high values indicate a rare term that occurs frequently in a given document) but very sparse, which affects the performance of machine learning algorithms. In contrast, word embeddings produced by representation learning algorithms are dense but do not provide direct interpretation.
Both approaches have a common pipeline—in the first step, they use an external algorithm to find potential concepts in a scientific text. Then, the suggestions are linked to the concepts using retrieval techniques such as dictionary search or vector spacing.
3.3. Representation Learning and fastText
Techniques for linking entities based on vector similarity can use either word embeddings or tf-idf vectors. The latter can be advantageous due to their dense vector structure and ability to be generated by powerful language models trained on large corpora. Neural networks are widely employed for their strong abilities in NLP (Natural Language Processing). The creation of ontologies relies heavily on text, suggesting the applicability of artificial neural networks (ANNs) in this context. The fastText [
37] is an algorithm based on representation learning that generates embeddings on the word level. A single-hidden layer neural network is trained to predict a word in context, and the learned word representations are then used as word embeddings.
3.4. Text Similarity Strategy for Matching Texts to Ontologies
Ontologies typically contain annotations for most of their classes and relations, potentially generating supervised datasets for machine-learning algorithms. However, we need to make several strong assumptions before employing a text similarity approach:
The distribution of texts provided as input is the same as the distribution of embeddings of annotation texts. This means that the input sentences should follow the same general structure, length, and vocabulary as ontology annotations to avoid prediction skewing for irrelevant reasons.
The best matching ontology is the one which provides annotations most similar to the input text. Since the considered methods are text-based, they will not rely on structures or hierarchies created by ontology classes and input text terms.
For below mentioned methods, we use fastText and BERT models. They were trained using texts from related domains, which will serve as a backbone for further processing. Following the notation introduced in
Section 3.1, we consider a “hard” mapping
directly to the set of ontologies of interest.
3.4.1. Zero-Shot Classification
The method is composed by assigning an ontology considering the similarity between annotation embeddings and input text embedding. This provides a simple method and does not require fine-tuning of the model, so a basis for further experimentation can be quickly established. Common similarity measures are cosine distances or Euclidean distances. In our experiments, we choose the first one because in some embedding algorithms the vector length can be affected by the size of the input text, so vectors corresponding to semantically similar texts generally point in the same direction, but are different in terms of the Euclidean distance.
3.4.2. Supervised Classification Based on Ontology Annotations
This approach relies on supervision provided by ontology annotation attributes of nodes and relations. Let us provide a set of ontologies, K. We can receive a dataset with pairs of annotations and the respective ontology source label and use it in supervised training. With the above assumptions, we can map the input texts to the ontologies by the trained model.
3.4.3. Negative Sampling
This approach is an extension of the above approach with the class “None”, which states that the input text is not to be assigned to any of the known ontologies. Thus, the annotation dataset is enhanced by:
Texts received from scientific papers from unrelated domains and labeled by the “None” class.
Sentences extracted from scientific papers from related knowledge domains with a different target during training. Instead of maximizing the model output scores for a ground truth class, the output scores for the “None” class are minimized for related input texts. This method is intended to partially compensate for the possible difference in input distribution between ontology annotations and scientific texts.
3.5. Text Processing
For the task of classifying scientific texts into the most relevant existing ontology, documents in Portable Document Files (PDFs) are used. One problem with PDFs is that they have an internal structure for printing by a physical printer device, and therefore keep metadata about the contained content with respect to its place on the paper. Therefore, it is complicated to extract the whole content of each paragraph. In case the file is parsed with the simple PDF tool and the line-break character is used to separate between paragraphs, only a single line is returned and not the entire paragraph. Another problem is related to documents with multiple columns of text on one page. In case the document does not contain a mark to the point where the paragraph continues, software tools for text extraction from PDF files usually continue with the nearest letter in the same line instead of going to the next line in the same column.
The found solution for retrieving text data from multi-column PDFs uses the engine contained in Microsoft Word. Its ability is able to pass both problems and extract the text without issues. It identifies meta-information in the content such as titles, paragraphs, and sentences. Some source documents contain irrelevant text to the topic of interest, such as references, acknowledgments, etc. The descriptions of ontologies are usually contained in Ontology Web Language (OWL) files. The OWL’s internal structure [
41] is a kind of XML structure for ontologies extended by RDF. Content describing classes and relationships can be marked by different tags depending on the ontology developer’s decision.
3.6. Pipeline for Comparing BERT and fastText Results
Before classifying articles according to the most relevant ontologies, we have in this context compared BERT and fastText. In this pipeline, we have used spaCy. It is a free, open-source library for advanced Natural Language Processing (NLP) in Python. The following text preprocessing pipeline for input embeddings is thus employed (the stars at the beginning of some points mark, which are applied to new sentences from scientific papers only):
- 1.
*Parse an input text into sentences by a SpaCy model.
- 2.
*Filter out invalid sentences, which contain less than two nouns or no verb.
- 3.
*Filter out sentences with non-paired parenthesis and ill-parsed formulas or composed terms.
- 4.
(BERT) Tokenize sentences with a tokenizer received from the model.
- 5.
(fastText) Transform to lowercase and split into words.
3.7. Classification of Output Embeddings
The output embeddings of the texts, obtained by BERT, are used as input data for classification algorithms, classifying a given input paragraph with respect to its relevance to the considered ontologies. The classification algorithms have been trained on the embeddings of the annotations of relations and classes from the considered ontologies because we know the ground truth (i.e., the ontology to which the annotation belongs) for them.
We have selected the following five classification algorithms implemented in scikit-learn [
42]:
Random Forest (RF): An ensemble classifier that fits a set of classification trees to different subsamples of the training dataset and uses an aggregation function to improve prediction accuracy and control overfitting. Typically, each tree in the ensemble is created using a surrogate sample (by bootstrapping algorithm—selection from source with repetition) from the training dataset. In addition, as each node is split during tree creation, the best split is found using all features from the input or a random subset of features of a given size. The goal is to reduce the variance of the forest estimator. The single decision tree typically has high variance and tends to overfit. The randomness introduced into the forests leads to decision trees with slightly decoupled prediction errors. By averaging these predictions, some errors can be mitigated. Random forests achieve lower variance by combining different trees, sometimes at the cost of a small increase in bias. Usually, the reduction in variance leads to an overall better model [
43].
Support Vector Machine (SVM): This is a classifier specifically designed to achieve the lowest possible prediction error, using a known relationship between the generalization error and the edge of the separating hyperplane. It uses only training points on both support hyperplanes of the boundary (support vectors), which leads to memory effectiveness. A simple SVM can only be used for linearly separable classes. For linearly non-separable classes, the data must first be transformed into linearly separable function sets in a high-dimensional vector space of functions using a suitable kernel. SVM classification has multi-class support, which is handled according to a one-versus-one or one-versus-rest scheme [
44].
Gaussian Process (GP): It was developed primarily for regression problems. A Gaussian Process Classifier (GPC) implements a collection of random variables indexed by a Euclidean space by placing a GP prior on latent functions. Its purpose is to provide a convenient formulation of the classification by a logistic link function. GPCs support multiclass classification by performing either one-vs.-rest or one-vs.-one training and prediction. A critical component of any GPC is the covariance function of the underlying GP. It encodes the assumptions about the similarity of Gaussian distributions corresponding to different points [
45].
K-Nearest Neighbors (k-NN): Neighbors-based classification stores individuals from the training dataset. A query instance is assigned to the class that has the most representatives within the point’s nearest neighbors. In nearest neighbor classification, uniform weights can be used, i.e., the value assigned to a query point is calculated by a simple majority vote of the nearest neighbors. In some cases, it is better to weigh the neighbors so that the nearest neighbors contribute more. For example, if the class of a queried point is computed from two nearest points and one of them is closer than the second by a defined metric, the resulting class in this case is the class of the closest point. The distance
d between two points can be calculated as:
, where
n is the dimension of the space and
, if
, this is the Manhattan distance and in the case of
, this is the Euclidean distance [
46].
Multilayer Perceptron (MLP): Given a set of features and targets, it can learn a nonlinear function approximator for classification or regression. Between the input and output layers, it can have one or more hidden layers. The input features are represented in the input layer consisting of a set of neurons. Using a weighted linear summation and a non-linear activation function, the neurons making up the hidden layer, transform the values from the previous layer. The last hidden layer then transfers its values to the output layer, which in turn transforms these values into output values. MLP has the advantage that it is able to learn nonlinear dependencies and also is capable of online learning, which allows learning models in real-time. However, a downside of the MLP with hidden layers is the non-convex loss function with multiple local minima. In the case of training MLP with the same data, the results may be different, when the MLP uses random weight initialization. A number of hyper-parameters are needed to be set for MLP such as the number of layers or the number of hidden neurons. MLPs also express sensitivity to feature scaling [
47].
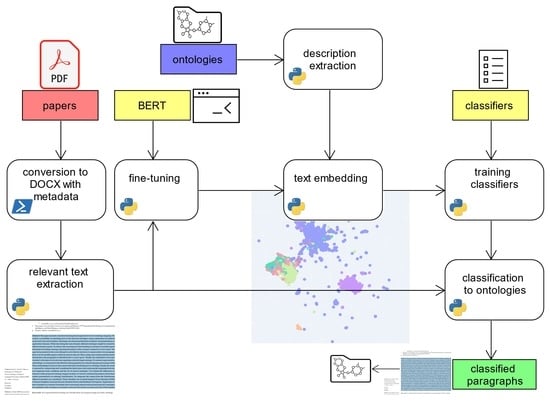
3.8. Summary of the Methodology
The summarization of the whole process classification is shown in
Figure 2. The core inputs are scientific papers in PDF and the output is a table of classified paragraphs. The table rows contain vectors with probabilities to which ontology each paragraph fits.
For this task, the relevant text first is extracted from the papers to obtain a dataset with the text of source paragraphs. As this poses the textual data, it is used for fine-tuning a BERT transformer for the later use of source text embeddings received from ontologies and papers.
In addition, descriptions of classes and relations are extracted from ontologies and stored as ontology description dataset. Then, the fine-tuned transformer is used to produce text embeddings of the descriptions. This yields a dataset containing the embeddings as a ground truth based on the text data from the descriptions obtained from ontologies.
The set then is used to train the classifiers of Random Forest, SVM, Gaussian Process, k-NN, and MLP. Those trained classifiers in turn are finally used together with the dataset with text paragraphs from papers for their classification regarding the best-fitting ontology.
4. Empirical Validation in Catalysis Research
A catalyst is a chemical that is not consumed in the course of a chemical reaction. The use of a catalyst in a chemical reaction usually allows the reaction to proceed more quickly and allows milder reaction conditions to prevail. Chemical synthesis based on catalysts is used in about 90 % of chemical processes in the chemical industry. The scientific field of catalysis is highly interconnected with other sciences and therefore spans many topics, from materials science to process design [
48,
49].
We perform our experiments using a set of five chemical domain ontologies (
Table 1) developed as part of the NFDI4Cat project [
2]. The National Cancer Institute Thesaurus (NCIT) [
50], Chemical Methods Ontology (CHMO) [
51], and Allotrope Foundation Ontology (AFO) [
52] ontologies are closely related to the chemical domain. Chemical Entities of Biological Interest (CHEBI) [
53] and Systems Biology Ontology (SBO) [
54], on the other hand, are expected to have a lesser connection to the chemical domain based on their names. This is not necessarily true for CHEBI, as it describes a plethora of chemical entities that are also relevant in the chemical domain, not just the biological domain. The SBO was chosen because it contains some general laboratory and computational contexts. It can also be observed as a kind of test to whether the tools used can also identify ontologies that do not match the text content.
4.1. Comparison of fastText and BERT
In this experiment, we have used 28 scientific papers composed of 25 research as well as 3 review papers for input to the evaluation. Those papers consist of 1.3 million symbols (1485 relevant paragraphs) and encompass the topic of the methanation of CO2. These articles have been extracted to Microsoft Word files by a PowerShell script. By Microsoft Word’s engine, paragraphs, and headings are recognized and marked properly, so paragraphs with relevant texts have been extracted by the template in Listing 1 and by the BERT embedding prepared for the final classification. In addition, these texts are used for fine-tuning BERT.
| Listing 1. Irrelevant parts in source text are titled by strings listed on the first line. Paragraphs in parts with different headings are stored and their headings are removed. |
![Computers 12 00014 i001]() |
We have selected the pretrained fastText model by [
55] and the
recobo/chemical-
bert-uncased [
56] checkpoint of a BERT implementation [
57] from the HuggingFace portal. In the preprocessing task, we use SpaCy [
58] with a ScispaCy [
59] model
en_ner_bc5cdr_md. For the remaining machine learning models, PyTorch implementations were used. For the visualization method Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) [
60], we used the implementation described in [
61].
Because we lack data with known ground truth, we evaluate the quality primarily by inspecting the resulting input sentence-annotation pairs.
4.1.1. Zero-Shot Setup
We start with representation learning of annotations using the fastText and BERT algorithms and examine the generated embeddings. For dimensionality reduction, we use the UMAP algorithm with a count of 15 neighbors, a minimum distance of 0.5, and a cosine metric. We found that with three-dimensional embeddings, much more information is preserved (and it is possible to split clusters that might be inseparable in 2D). The results are shown in
Figure 3. Blue dots denote embeddings from NCIT, red dots embeddings from CHMO ontology, green dots embeddings from AFO, purple dots embeddings from SBO, and orange dots represent CHEBI ontology embeddings. Three example sentences with the annotations assigned to them by fastText and BERT are shown in
Table 2. The annotation “carbon dioxide” was assigned by BERT to all three new example sentences.
4.1.2. Ontology Matching as Text Classification
As mentioned in
Section 3.4, another possible strategy to solve the problem is to treat it as a classification task into multiple classes. If the distributions of input texts and corresponding ontologies are the same, we can train a classifier on descriptions of ontology classes and relations and use them as input texts.
We have implemented this using embedding ontology annotations by BERT and training a dense (fully connected) 768-dimensional hidden layer over them. Minority data points are proportionally overestimated due to the significant size differences between ontologies. The classifier achieves a validation accuracy of 0.987 after a single-shot validation of the annotations of all nodes, indicating a good separability for different ontologies, cf.
Figure 4 and
Figure 5.
Figure 4.
Training plot of the ontology classifier model and a 3-dimensional projection of the BERT embeddings before tuning. The visualization provides an intuition about the distribution gap between ontology annotation and input texts, for which we are looking for the most relevant ontology. (a) Training and validation (blue resp. yellow) accuracy dynamics during the training. (b) BERT embeddings of ontology annotations and text sentences extracted from 28 scientific papers.
Figure 4.
Training plot of the ontology classifier model and a 3-dimensional projection of the BERT embeddings before tuning. The visualization provides an intuition about the distribution gap between ontology annotation and input texts, for which we are looking for the most relevant ontology. (a) Training and validation (blue resp. yellow) accuracy dynamics during the training. (b) BERT embeddings of ontology annotations and text sentences extracted from 28 scientific papers.
Figure 5.
Visualization of the BERT embeddings along with the classifier MLP hidden states in the ontology classification task. The model on the right takes as input vectors produced by the untuned BERT, visualized in the
Figure 4b. (
a) Negative sampling fine-tuned BERT embeddings, a 2d-image of the original space projected down and visualized in the 3 dimensions. (
b) Hidden layer outputs of the classifier model on top of the BERT, a 2d-image of the hidden activation space projected down and visualized in the 3 dimensions.
Figure 5.
Visualization of the BERT embeddings along with the classifier MLP hidden states in the ontology classification task. The model on the right takes as input vectors produced by the untuned BERT, visualized in the
Figure 4b. (
a) Negative sampling fine-tuned BERT embeddings, a 2d-image of the original space projected down and visualized in the 3 dimensions. (
b) Hidden layer outputs of the classifier model on top of the BERT, a 2d-image of the hidden activation space projected down and visualized in the 3 dimensions.
Those visualizations and
Table 3 allow to suppose that the model embeds input papers separately from ontology annotations, which may indicate a distribution shift between annotations and sentences.
However, when we have preprocessed the input texts and embedded them, our inspection has demonstrated that their distribution differs significantly in comparison to the distribution of the ontology annotations. The projection in
Figure 4 and
Figure 5 show a dense separated group of texts parsed from scientific papers.
While the BERT embeddings are more discriminative for the ontology classification task, the assigned sentences and the low-dimensional embeddings in
Figure 5 show that this approach is more sensitive to the distributional shift problem.
4.1.3. Negative Sampling
To address this problem, we included scientific texts in the training dataset. We selected 400 texts from the area of chemical science (as positive examples) and 400 from non-related areas (as negative examples). The model has two targets during training:
Cross-entropy loss when the input is an ontology annotation (as before).
Binary cross-entropy loss when the input is a text from a scientific paper. The model minimizes the probability of outputting a special class “Negative” for a related scientific text and maximizes it for non-related ones.
In our setup, we first train the head over BERT to convergence and leave the backbone frozen. If we consider only the ontology annotations and leave aside the example sentences, the model achieves a validation accuracy of 0.984; this value is similar to the accuracy of the classifier described before.
We then fine-tune the whole BERT model. It achieves a validation accuracy of 0.958 after single-shot validation on the combined annotation and paper sentence dataset, with the confusion matrix shown in
Figure 6, where values close to one are denoted blue and values close to zero denoted in the white color. By mixing sentences from irrelevant and relevant scientific texts, the classification accuracy was improved compared to the BERT classifier, as shown later.
Despite the good separability of the individual ontologies and the additional optimization criterion, the UMAP embeddings are similar to the previous setting in terms of clustering the input sets in a separate subspace.
It is worth noting that the classifier and negative sampling models produce softmax scores that can be interpreted as class probability distributions. However, neural networks tend to be overconfident in their results [
62], so additional calibration is required before using the results for the relevance decision.
4.1.4. Statistical Results
We compare the models using the Friedman test first, which checks whether the models have the same performance. We make a stratified split of the validation dataset with the ontology annotations into fifty samples and test the following null hypothesis:
Hypothesis 1. All six models perform the same on the validation splits.
The Friedman test has rejected the null hypothesis on the significance level of 5%. For the post-hoc analysis, we have employed the Wilcoxon signed-rank test with a two-sided alternative for all pairs of the compared results, because of the inconsistency of the more common mean ranks post hoc test, as pointed out in [
63]. For correction to multiple hypotheses testing, we have used the Holm method. We make the following assumptions about the algorithms, which will imply the alternative hypotheses for the null hypotheses 2–4 discussed below:
For a larger k, the k-NN classifier can work the same or better than the 1-NN.
The neural network model can fit training data the same or better than the k-NN.
The negative sampling results in a non-decrease or an improvement in the model generalization.
Hypothesis 2. [Null for k-NN models] The 10-NN models perform the same as their 1-NN variants.
While the 1-NN is a common setting for many NLP systems, it may produce complex decision boundaries and lead to overfitting. We test a larger k versus one to determine if this is an issue in our setup.
Hypothesis 3. [Null for neural network classifier] The neural-network model performs the same as the k-NN model on BERT embeddings and on fastText embeddings.
The assumption behind this hypothesis is that a neural network as a universal approximator can fit data better than a nearest-neighbor classifier.
Hypothesis 4. [Null for the fine-tuned model with negative sampling] For each of the considered models: The fine-tuned BERT with negative sampling performs the same as other considered models.
Our assumption is that additional sampled sentences will improve the model’s performance and will help to avoid overfitting when the model is fine-tuned as a whole instead of its head only.
Hypothesis 5. [Null for the rest] For each remaining pair: Both models from the pair have the same performance.
We indicate the relative model performance in
Figure 7, where high values are denoted in blue and low values in white color.
The test rejected the null hypotheses on the 5% significance level, except for Hypothesis 2. This hypothesis was rejected only for the fastText embeddings, which might be explained by a relatively sharp boundary between individual classes on UMAP visualizations of the embeddings. If this holds true for the original space, larger k may suppress outlier noise while decreasing the classification accuracy near it.
4.2. Experimental Setup and Preprocessing for the Comparison of Classifiers
In the first part, the received portable document files were converted to Microsoft Word file format using PowerShell scripts. The transformed files were processed using a Python library for parsing docx file format and SpaCy. As a result, the considered paragraphs were extracted for classification with respect to the most relevant ontology 1. We skipped paragraphs containing acknowledgments, references, titles, and paragraphs that were too short (shorter than 170 letters).
The extracted paragraphs were also used for fine-tuning the BERT model. The chosen pretrained version of BERT was recobo/chemical-bert-uncased from the Huggingface portal [
56]. The fine-tuning is located in the Listing 2. Using the fine-tuned BERT, each paragraph was converted to a 768-dimensional numerical vector. The annotations in the specifications of the given ontologies were extracted using the XML parser for Python called
BeautifulSoup. The parsing code can be found in the Listing 3. The individual annotations were again embedded into the 768-dimensional vector space. For this, the fine-tuned BERT presented in the previous section is used.
| Listing 3. Tags that stored relevant descriptions are in the list named ‘contentTags’. Because the OWL contains name-spaces owl, rdf, xml, rdfs, and obo, they had to be loaded. The output is one text file per ontology file, where each row contains one node or relation description. |
![Computers 12 00014 i003]() |
The classifiers were chosen from ScikitLearn [
42]. Each classifier has many hyperparameters. The optimal value of each hyperparameter was determined using a cross-validation algorithm with 5 folds. It was applied with a grid search to choose the optimal values from the given set. These values are listed in
Table 4. In order to mitigate overfitting, training data from large ontologies were under-sampled. For the statistic computations, we have used the pingouin, scipy, and statsmodels python’s libraries.
| Listing 2. The selected pretrained version of the BERT from the Huggingface portal was recobo/chemical-bert-uncased [56]. The model is trained using a CUDA device in an ideal environment, which makes tuning significantly faster than on the CPU. The source data are stored in a variable dataset. The used optimizer is Adaptive Moment Estimation. |
![Computers 12 00014 i002]() |
4.3. Comparison of Important Classifiers on Considered Ontologies
The classes to which classifiers assign new parts of text are ontologies from
Table 1. The source dataset has been split into training and testing datasets in stratified proportion 1:1. The data in the training dataset originating from large ontologies are unbalanced. We needed to mitigate overfitting during the training part. We have decided to under-sample the training dataset parts. The testing dataset has been split into 20 disjoint subsets, assuming that disjointness is a sufficient condition for their independence.
Aggregated statistic results of the predictive accuracy in the classification of all 20 testing datasets are in
Table 5. The table is enhanced with boxplots in
Figure 8, which presents the following quality measures for each classifier: accuracy, F1 score, precision, and recall. The RF classifier had the worst results of all classifiers. Other classifiers had much better statistical results. The Gaussian process had the best accuracy, its mean accuracy was 97% and it had a very low standard deviation.
Differences between the considered classifiers were tested for significance using the Friedman test. The basic null hypothesis that the average accuracy of all five classifiers is identical was strongly rejected with a significance level of
. The results of the Wilcoxon test are shown in
Table 6, with the best results for the SVM classifier and the Gaussian process classifier.
4.4. Classification of Scientific Texts with Respect to Relevant Ontologies
In the following experiment, we did not have a ground truth about which of the available ontologies was most relevant for each text passage considered. We used two collections of scientific papers from the field of chemical catalysis research. The small collection is the same as in the previous experiment and the large collection is the digital archive of papers from Leibniz Institute for Catalysis scientists (except for very few papers with read-only permission). This set of documents contains 3450 portable document files. We extracted 144,490 relevant paragraphs from them. The BERT embeddings of these paragraphs were classified by the five classifiers. Each paragraph’s embedding can be classified into more than one target class with non-zero probability. The probability distribution over all classes is used as the confidence of classification into each of them.
4.4.1. Results for the Small Dataset
Figure 9 shows counts of paragraphs each classifier assigned to each ontology. While the AFO is represented as blue bars, the CHEBI is shown in orange, the CHMO in green, the NCIT is represented as red bars, and the SBO is shown in purple. The Gaussian process, the k-NN, the MLP, and the SVM classifier assigned almost all paragraphs to the NCIT ontology. The RF classifier is the most uncertain of all the classifiers and assigned most of the paragraphs to the CHEBI ontology, but some paragraphs to each of the other four ontologies.
Figure 10 shows the sum of the confidences of the class assignment. The confidence of the MLP and the SVM is very high, while the confidence of the Random Forest and the Gaussian process is much lower. The k-NN classifier also has fairly high confidence.
Figure 11 shows the spread between the predicted ontology confidence and the second-highest class confidence. Even here, the highest values are obtained by SVM and MLP, while GP and RF have only a small difference between the predicted and the second-highest ontology confidence. The k-NN has a quite high difference, but not as high as MLP or SVM.
4.4.2. Results for the Large Dataset
Figure 12 shows the count of assignment of paragraphs from the second dataset to ontology by each classifier with coloring similar to the charts presented in the experiment with the small dataset. The GP, k-NN, MLP, and SVM have assigned almost all paragraphs to the NCIT ontology. The RF has the most unstable predictions of all the classifiers. It has assigned most of the paragraphs to the CHMO ontology but also assigned some paragraphs to the other four ontologies.
Figure 13 using the confidence of the class predictions shows that the confidence of the SVM is very high, while that of the GP and the RF is much lower. The MLP and the k-NN classifier also have fairly high confidence.
Figure 14 shows the spread between the predicted ontology confidence and the second-highest class confidence. The results are similar to those in the first experiment; the highest values are obtained by the SVM, while the GP and RF have only a small difference between the predicted and second-highest ontology confidence. The MLP and k-NN have quite a high margin, but not as high as the SVM.
4.4.3. Summary of Results for Both Datasets
This experiment has returned results, from which we can say that SVM has good accuracy in testing data according to all considered quality measures. The results of the confidence margins between the top two most relevant ontologies and of the predicted most relevant ontology confidence are the best of all considered classifiers. Hence, the results indicate that for a large majority of the unknown scientific texts, the most relevant ontology is NCIT.
5. Discussion and Outlook
Ontologies provide a way to express knowledge and data in a formalized and standardized way. However, since creating and maintaining an ontology is a manual process, there are different conceptualizations and different representations of knowledge. This poses a challenge when a researcher wants to select the most appropriate ontology for a particular scientific text (i.e., his/her research). In this work, we investigate the possibility of automatically selecting the most relevant ontologies for given scientific texts in order to reduce the workload for domain experts. We are not aware of other works on this topic, so we are not able to discuss them and compare the proposed approach with previous methods.
In the research conducted, it was found that the distributions of scientific texts differ significantly from those in ontology annotations. Despite a good separability of the considered ontology annotations from each other, as well as the sets of positively and negatively valued articles, the investigated approach leads to a mapping in separate subsets of the embedding space. This is true also for the most sophisticated of the three settings studied, where BERT was fine-tuned with both ontology annotations and scientific papers from related and unrelated areas.
To mitigate such inadequate coverage of unknown scientific texts, the following research could contain an intermediate step recognition of entities. The use of recognized entities instead of original texts may lead to separating the information in scientific texts directly related to terms from ontologies from unrelated terms, phrases, and other partitions of the text that were not eliminated by preprocessing.
Furthermore, we report on the use of classifiers in combination with representation learning through the fine-tuned BERT. We used the embedding of each paragraph from PDFs as input to the classifiers. We made tests using five different classifiers, specifically the Gaussian Process, k-NN, Multilayer Perceptron, Random Forest, and Support Vector Machine. The Random Forest was the worst of all considered classifiers; its precision as well as other quality measures were the lowest among all classifiers. The best results were obtained by the Gaussian Process and the Support Vector Machine.
In the last test, the used classifiers were compared using scientific papers from the field of catalysis. The ground truth of the papers was not known. The k-NN and the Gaussian Process had very small ranges between the predicated and the second highest confidence. The Support Vector Machine had the highest confidence of all tested classifiers. It also had the highest margin among them.
Since there is no ground truth for the classification of the scientific articles, it is hard to assess this experiment. Hence, we want to test methods that reduce the impact of the unknown ground truth. Our idea is to use interpolation between annotations using public alternatives to GPT-2 and GPT-3 networks that are available to use in local code. When the GPT-4 will release, we want to try this transformer as well. GPT (Generative Pre-trained Transformer) [
64] stands for a set of pre-trained language models developed by the OpenAI group. The transformers can be used to tackle specific language-related tasks, as they have been trained with a large dataset of textual information. The BERT has been trained with book data and wiki data containing over 3.3 billion tokens. Thus, BERT is popular in natural language understanding tasks such as text classification. However, BERT, being a masked language model, can only learn a contextual representation of words. Therefore, it cannot organize or generate language, rendering it unsuitable for concept-generation tasks. The GPT is an autoregressive language model that is trained to predict the next word based on all previous words.
In the following experiments, it is desirable to try more different transformers. We want also to gain information from ANNs related to learning. The main direction of our research is to extend and integrate existing ontologies. Our plan contains also the usage of graph neural networks (GNNs) to apply them in the representation learning of ontologies.
Author Contributions
Conceptualization, M.H., A.S.B. and N.K.; data resources, M.H. and A.S.B.; investigation, L.K. and U.Y.; methodology, M.H., U.Y. and L.K.; software, L.K. and U.Y.; supervision, M.H.; writing—original draft preparation, L.K.; writing—review and editing, M.H., U.Y., A.S.B. and N.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been supported by the Grant Agency of the Czech Technical University in Prague, grant No. SGS20/208/OHK3/3T/18, and by the German Research Foundation (DFG) funded projects NFDI2/12020 and 467401796.
Data Availability Statement
The data used in the experimental evaluation in
Section 4 are available upon request from the authors.
Acknowledgments
We appreciate the very extensive and useful feedback that we received from the five anonymous reviewers of the predecessor version of the paper, presented at the 10th International Workshop on Computational Intelligence and Data Mining.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AFO | Allotrope Foundation Ontology |
| ANN | Artificial Neural Network |
| ART | Adaptive Resonance Theory |
| BERT | Bidirectional Encoder Representations from Transformers |
| BiLSTM | Bidirectional Long Short-Term Memory |
| CNN | Convolutional Neural Network |
| FAIR | Findable, Accessible, Interoperable and Reusable |
| FN | False Negative |
| FP | False Positive |
| GNN | Graph Neural Network |
| GP | Gaussian Process |
| GPC | Gaussian Process Classifier |
| GPT | Generative Pre-Trained Transformer |
| GRU | Gated Recurrent Unit |
| CHEBI | Chemical Entities of Biological Interest |
| CHMO | Chemical Methods Ontology |
| KNN | K Nearest Neighbors |
| LSTM | Long Short-Term Memory |
| MLP | Multilayer Perceptron |
| NCBO | National Center for Biomedical Ontology |
| NCIT | National Cancer Institute Thesaurus |
| OWL | Web Ontology Language |
| PDF | Portable Document File |
| RDF | Resource Description Framework |
| RF | Random Forest |
| SBO | System Biology Ontology |
| SVM | Support Vector Machine |
| TF-IDF | Term Frequency – Inverse Document Frequency |
| TN | True Negative |
| TP | True Positive |
| XML | Extensible Markup Language |
References
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wulf, C.; Beller, M.; Boenisch, T.; Deutschmann, O.; Hanf, S.; Kockmann, N.; Kraehnert, R.; Oezaslan, M.; Palkovits, S.; Schimmler, S.; et al. A Unified Research Data Infrastructure for Catalysis Research—Challenges and Concepts. ChemCatChem 2021, 13, 3223–3236. [Google Scholar] [CrossRef]
- Gruber, T.R. A translation approach to portable ontology specifications. Knowl. Acquis. 1993, 5, 199–220. [Google Scholar] [CrossRef]
- Grühn, J.; Behr, A.S.; Eroglu, T.H.; Trögel, V.; Rosenthal, K.; Kockmann, N. From Coiled Flow Inverter to Stirred Tank Reactor—Bioprocess Development and Ontology Design. Chem. Ing. Tech. 2022, 94, 852–863. [Google Scholar] [CrossRef]
- Horsch, M.; Petrenko, T.; Kushnarenko, V.; Schembera, B.; Wentzel, B.; Behr, A.; Kockmann, N.; Schimmler, S.; Bönisch, T. Interoperability and Architecture Requirements Analysis and Metadata Standardization for a Research Data Infrastructure in Catalysis. In Proceedings of the Data Analytics and Management in Data Intensive Domains; Pozanenko, A., Stupnikov, S., Thalheim, B., Mendez, E., Kiselyova, N., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 166–177. [Google Scholar] [CrossRef]
- Fensel, D. Ontologies: A Silver Bullet for Knowledge Management and Electronic Commerce, 2nd ed.; Springer: Berlin/Heidelberg, Germany; London, UK, 2011. [Google Scholar] [CrossRef]
- Guarino, N. Formal Ontology and Information Systems. In Proceedings of the FOIS’98 Conference, Trento, Italy, 6–8 June 1998; IOS Press: Amsterdam, The Netherlands, 1998; pp. 3–15. [Google Scholar]
- Martínez-Romero, M.; Jonquet, C.; O’Connor, M.J.; Graybeal, J.; Pazos, A.; Musen, M.A. NCBO Ontology Recommender 2.0: An enhanced approach for biomedical ontology recommendation. J. Biomed. Semant. 2017, 8, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Aswadi, F.; Chan, H.; Gan, K. Extracting Semantic Concepts and Relations from Scientific Publications by Using Deep Learning. In Proceedings of the IRICT 2020, Langkawi, Malaysia, 21–22 December 2021; pp. 374–383. [Google Scholar]
- Gupta, N.; Podder, S.; Annervaz, K.; Sengupta, S. Domain Ontology Induction Using Word Embeddings. In Proceedings of the ICMLA, Anaheim, CA, USA, 18–20 December 2016; pp. 115–119. [Google Scholar]
- Katyshev, A.; Anikin, A.; Denisov, M.; Petrova, T. Intelligent Approaches for the Automated Domain Ontology Extraction. In Proceedings of the International Congress on Information and Communication Technology, London, UK, 25–26 February 2021; pp. 410–417. [Google Scholar]
- Althubaiti, S.; Kafkas, S.; Abdelhakim, M.; Hoehndorf, R. Combining Lexical and Context Features for Automatic Ontology Extension. J. Biomed. Semant. 2020, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Espinoza-Anke, L.; Ronzano, F.; Saggion, H. Hypernym Extraction: Combining Machine-Learning and Dependency Grammar. In Proceedings of the CICLing, Cairo, Egypt, 14–20 April 2015; pp. 372–383. [Google Scholar]
- Martel, F.; Zouaq, A. Taxonomy Extraction Using Knowledge Graph Embeddings and Hierarchical Clustering. In Proceedings of the SAC’21, Virtual, 22–26 March 2021; pp. 836–844. [Google Scholar]
- Navarro-Almanza, R.; Juárez-Ramírez, R.; Licea, G.; Castro, J.R. Automated Ontology Extraction from Unstructured Texts using Deep Learning. In Intuitionistic and Type-2 Fuzzy Logic Enhancements in Neural and Optimization Algorithms: Theory and Applications; Springer: Berlin/Heidelberg, Germany, 2020; pp. 727–755. [Google Scholar]
- Bento, A.; Zouaq, A.; Gagnon, M. Ontology Matching Using Convolutional Neural Networks. In Proceedings of the LREC, Marseille, France, 11–16 May 2020; pp. 5648–5653. [Google Scholar]
- Chakraborty, J.; Yaman, B.; Virgili, L.; Konar, K.; Bansal, S. OntoConnect: Results for OAEI 2020. In Proceedings of the OM ISWC, Virtual, 2 November 2020; pp. 204–210. [Google Scholar]
- Hao, J.; Lei, C.; Efthymiou, V.; Quamar, A.; Özcan, F.; Sun, Y.; Wang, W. MEDTO: Medical Data to Ontology Matching Using Hybrid Graph Neural Networks. In Proceedings of the KDD’21, Virtual, 14–18 August 2021; pp. 2946–2954. [Google Scholar]
- Wu, J.; Lv, J.; Guo, H.; Ma, S. Daeom: A Deep Attentional Embedding Approach for Biomedical Ontology Matching. Appl. Sci. 2020, 10, 909. [Google Scholar] [CrossRef] [Green Version]
- Hourali, M.; Montazer, G. Using ART2 Neural Network and Bayesian Network for Automating the Ontology Constructing Process. Procedia Eng. 2012, 29, 3914–3923. [Google Scholar] [CrossRef]
- Mercier, C.; Chateau-Laurent, H.; Alexandre, F.; Viéville, T. Ontology as Neuronal-Space Manifold: Towards Symbolic and Numerical Artificial Embedding. In Proceedings of the Workshop on Knowledge Representation for Hybrid and Compositional AI, Virtual, 8–9 February 2021; pp. 1–11. [Google Scholar]
- Kolozali, S.; Fazekas, G.; Barthet, M.; Sandler, M. A Framework for Automatic Ontology Generation Based on Semantic Audio analysis. In Proceedings of the Audio Engineering Society International Conference, Los Angeles, CA, USA, 9–12 October 2014; pp. 87–96. [Google Scholar]
- Li, G. CNN Based Ontology Learning Algorithm and Applied in PE Data. IAENG Int. J. Comput. Sci. 2021, 48, 1–8. [Google Scholar]
- Mueller, R.; Abdullaev, S. Deep Cause: Hypothesis Extraction from Information Systems Papers with Deep Learning for Theory Ontology Learning. In Proceedings of the Annual Hawaii International Conference on System Sciences, Maui, HI, USA, 8–11 January 2019; pp. 6250–6259. [Google Scholar]
- Petrucci, G.; Rospocher, M.; Ghindini, C. Expressive Ontology Learning as Neural Machine Translation. J. Web Semant. 2018, 52–53, 66–82. [Google Scholar] [CrossRef]
- Potoniec, J. Learning OWL 2 Property Characteristics as an Explanation for an RNN. Bull. Pol. Acad. Sci. Tech. Sci. 2020, 68, 1481–1490. [Google Scholar]
- Memariani, A.; Glauer, M.; Neuhaus, F.; Mossakowski, T.; Hatings, J. Automated and Explainable Ontology Extension Based on Deep Learning: A Case Study in the Chemical Domain. In Proceedings of the 3rd International Workshop on Data Meets Applied Ontologies, Hersonissos, Greece, 29 May–2 June 2021; pp. 1–16. [Google Scholar]
- Oba, A.; Paik, I.; Kuwana, A. Automatic Classification for Ontology Generation by Pretrained Language Model. In Proceedings of the International Conference on Industrial, Engineering and Other Applications of Applied Intelligent Systems, London, UK, 7–9 July 2021; pp. 210–221. [Google Scholar]
- Teslya, N.; Savosin, S. Matching Ontologies with Word2Vec-Based Neural Network. In Proceedings of the ICCSA, Saint Petersburg, Russia, 1–4 July 2019; pp. 745–756. [Google Scholar]
- Ristoski, P.; Paulheim, H. Rdf2vec: Rdf Graph Embeddings for Data Mining. In Proceedings of the International Semantic Web Conference, Kobe, Japan, 17–21 October 2016; pp. 498–514. [Google Scholar]
- Ritchie, A.; Chen, J.; Castro, L.; Rebholz-Schuhmann, D.; Jiménez-Ruiz, E. Ontology Clustering with OWL2Vec. In Proceedings of the DeepOntoNLP, Hersonissos, Greece, 6–10 June 2021; pp. 54–61. [Google Scholar]
- Petrucci, G.; Ghindini, C.; Rospocher, M. Ontology Learning in the Deep. In Proceedings of the EKAW, Bologna, Italy, 19–23 November 2016; pp. 480–495. [Google Scholar]
- Hirschman, L.; Krallinger, M.; Valencia, A.; Fluck, J.; Mevissen, H.T.; Dach, H.; Oster, M.; Hofmann-Apitius, M. ProMiner: Recognition of Human Gene and Protein Names using regularly updated Dictionaries. In Proceedings of the Second BioCreAtIvE Challenge Evaluation Workshop, Madrid, Spain, 23–25 April 2007; pp. 149–151. [Google Scholar]
- Morgan, A.A.; Lu, Z.; Wang, X.; Cohen, A.M.; Fluck, J.; Ruch, P.; Divoli, A.; Fundel, K.; Leaman, R.; Hakenberg, J.; et al. Overview of BioCreative II gene normalization. Genome Biol. 2008, 9 (Suppl. S2), S3. [Google Scholar] [CrossRef] [PubMed]
- Leaman, R.; Islamaj Dogan, R.; Lu, Z. DNorm: Disease name normalization with pairwise learning to rank. Bioinformatics 2013, 29, 2909–2917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karadeniz, İ.; Özgür, A. Linking entities through an ontology using word embeddings and syntactic re-ranking. BMC Bioinform. 2019, 20, 156. [Google Scholar] [CrossRef] [Green Version]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching Word Vectors with Subword Information. arXiv 2016, arXiv:1607.04606. [Google Scholar] [CrossRef] [Green Version]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers); Association for Computational Linguistics: Minneapolis, MN, USA, 2019; pp. 4171–4186. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, F.; Hu, Y.; Shi, C.; Fung, P. NER-BERT: A Pre-trained Model for Low-Resource Entity Tagging. CoRR 2021, arXiv:2112.00405. [Google Scholar]
- Lu, K.; Grover, A.; Abbeel, P.; Mordatch, I. Pretrained Transformers as Universal Computation Engines. CoRR 2021, arXiv:2103.05247. [Google Scholar] [CrossRef]
- Group, O.W. OWL. 2012. Available online: https://www.w3.org/OWL/ (accessed on 4 January 2023).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ho, T.K. Random decision forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, pp. 278–282. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.J.; Bach, F. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Rasmussen, C.E.; Williams, C.K.I. Gaussian Processes for Machine Learning (Adaptive Computation and Machine Learning); The MIT Press: Cambridge, MA, USA, 2005; pp. 33–77. [Google Scholar]
- Kramer, O. K-Nearest Neighbors. In Dimensionality Reduction with Unsupervised Nearest Neighbors; Springer: Berlin/Heidelberg, Germany, 2013; pp. 13–23. [Google Scholar] [CrossRef]
- Vang-Mata, R. Multilayer Perceptrons: Theory and Applications; Computer Science, Technology and Applications Series; Nova Science Publishers: Hauppauge, NY, USA, 2020. [Google Scholar]
- Benvenuto, M.; Plauman, H. Industrial Catalysis; De Gruyter STEM, De Gruyter: Berlin, Germany; Boston, MA, USA, 2021. [Google Scholar]
- Weiner, S.C. Technology vision 2020: The U.S. chemical industry. In Air Pollution in the 21st Century; Studies in Environmental Science; Schneider, T., Ed.; Elsevier: Amsterdam, The Netherlands, 1998; Volume 72, pp. 915–921. [Google Scholar] [CrossRef]
- National Cancer Institue. National Cancer Institue Thesaurus, 2022. Available online: https://ncit.nci.nih.gov (accessed on 1 December 2021).
- Batchelor, C. Chemical Methods Ontology. 2022. Available online: http://purl.obolibrary.org/obo/chmo.owl (accessed on 1 December 2021).
- Allotrope Foundation. Allotrope Foundation Ontology, 2022. Available online: https://www.allotrope.org/ontologies (accessed on 1 December 2021).
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2015, 44, D1214–D1219. [Google Scholar] [CrossRef]
- Nguen, T.; Karr, J.; Sheriff, R. Systems Biology Ontology. 2022. Available online: http://biomodels.net/SBO/ (accessed on 12 December 2022).
- Kim, E.; Jensen, Z.; van Grootel, A.; Huang, K.; Staib, M.; Mysore, S.; Chang, H.S.; Strubell, E.; McCallum, A.; Jegelka, S.; et al. Inorganic Materials Synthesis Planning with Literature-Trained Neural Networks. J. Chem. Inf. Model. 2020, 60, 1194–1201. [Google Scholar] [CrossRef] [Green Version]
- Company, R.A. BERT for Chemical Industry. 2022. Available online: https://huggingface.co/recobo/chemical-bert-uncased (accessed on 21 November 2022).
- Hugging Face. BERT. 2022. Available online: https://huggingface.co/docs/transformers/model_doc/bert (accessed on 21 November 2022).
- Honnibal, M.; Montani, I. SpaCy 2: Natural Language Understanding with Bloom Embeddings, Convolutional Neural Networks and Incremental Parsing. Available online: https://spacy.io/ (accessed on 21 November 2022).
- Neumann, M.; King, D.; Beltagy, I.; Ammar, W. ScispaCy: Fast and Robust Models for Biomedical Natural Language Processing. In Proceedings of the 18th BioNLP Workshop and Shared Task; Association for Computational Linguistics: Florence, Italy, 2019; pp. 319–327. [Google Scholar] [CrossRef] [Green Version]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Grossberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Gal, Y. Uncertainty in Deep Learning. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2016. [Google Scholar]
- Benavoli, A.; Corani, G.; Mangili, F. Should We Really Use Post-Hoc Tests Based on Mean-Ranks? J. Mach. Learn. Res. 2016, 17, 1–10. [Google Scholar]
- Zhu, Q.; Luo, J. Generative Pre-Trained Transformer for Design Concept Generation: An Exploration. Proc. Des. Soc. 2022, 2, 1825–1834. [Google Scholar] [CrossRef]
Figure 1.
BERT (Bidirectional Encoder Representations from Transformers) architecture [
38] the figure is reproduced under the CC-BY licese. An input sequence of words is divided into tokens and each token is encoded as a vector. There is one numeric vector per token in the BERT’s outputs. The output vector marked by C is subsequently used for classification.
Figure 1.
BERT (Bidirectional Encoder Representations from Transformers) architecture [
38] the figure is reproduced under the CC-BY licese. An input sequence of words is divided into tokens and each token is encoded as a vector. There is one numeric vector per token in the BERT’s outputs. The output vector marked by C is subsequently used for classification.
Figure 2.
Process diagram of the whole methodology.
Figure 2.
Process diagram of the whole methodology.
Figure 3.
Annotation embeddings produced by fastText and BERT, a 2d-image of the embeddings projected down to 3 dimensions. In the case of fastText, SBO, AFO, and CHMO annotations are located in tiny areas, primarily close to the center of the image. Note that fastText struggles to group smaller ontologies into more dense and decoupled clusters, while BERT provides embeddings with better separability of annotations of individual ontologies.
Figure 3.
Annotation embeddings produced by fastText and BERT, a 2d-image of the embeddings projected down to 3 dimensions. In the case of fastText, SBO, AFO, and CHMO annotations are located in tiny areas, primarily close to the center of the image. Note that fastText struggles to group smaller ontologies into more dense and decoupled clusters, while BERT provides embeddings with better separability of annotations of individual ontologies.
Figure 6.
Confusion matrix of the MLP classification over fine-tuned BERT for a dataset consisting of the annotations from all five considered ontologies and the sentences of the additional 400 related and 400 unrelated scientific papers. An additional column to the right is related to the training set containing an additional label “Positive”, which is the opposite of the “Negative” label, and is being used during training.
Figure 6.
Confusion matrix of the MLP classification over fine-tuned BERT for a dataset consisting of the annotations from all five considered ontologies and the sentences of the additional 400 related and 400 unrelated scientific papers. An additional column to the right is related to the training set containing an additional label “Positive”, which is the opposite of the “Negative” label, and is being used during training.
Figure 7.
The comparison matrix of the six considered models. The -th element indicates a number of splits where the i-th model performed better than j-th. Except for the one- and ten-nearest-neighbor over BERT embeddings, all the models demonstrate statistically significant differences. BERT NN denotes a neural network classifier trained over BERT embeddings.
Figure 7.
The comparison matrix of the six considered models. The -th element indicates a number of splits where the i-th model performed better than j-th. Except for the one- and ten-nearest-neighbor over BERT embeddings, all the models demonstrate statistically significant differences. BERT NN denotes a neural network classifier trained over BERT embeddings.
Figure 8.
The quality measures with a standard deviation of the considered classifiers on testing datasets. For each classifier was computed Accuracy, F1 score, Precision, and Recall. The worst result has the Random forest classifier, the others are significantly better.
Figure 8.
The quality measures with a standard deviation of the considered classifiers on testing datasets. For each classifier was computed Accuracy, F1 score, Precision, and Recall. The worst result has the Random forest classifier, the others are significantly better.
Figure 9.
Number of paragraphs of the collection of 28 scientific papers regarding the topic of methanation of CO2 predicted by highest confidence to target class based on five ontologies.
Figure 9.
Number of paragraphs of the collection of 28 scientific papers regarding the topic of methanation of CO2 predicted by highest confidence to target class based on five ontologies.
Figure 10.
Sum of prediction confidences with regards to the five ontologies for the collection of 28 scientific papers regarding the topic of methanation of CO2.
Figure 10.
Sum of prediction confidences with regards to the five ontologies for the collection of 28 scientific papers regarding the topic of methanation of CO2.
Figure 11.
Sum of margins between top two confidences for the collection of 28 scientific papers regarding the topic of methanation of CO2.
Figure 11.
Sum of margins between top two confidences for the collection of 28 scientific papers regarding the topic of methanation of CO2.
Figure 12.
Counts of paragraphs of the collection of scientific papers from the digital archive of papers from Leibniz Institute for Catalysis predicted by highest confidence to target class.
Figure 12.
Counts of paragraphs of the collection of scientific papers from the digital archive of papers from Leibniz Institute for Catalysis predicted by highest confidence to target class.
Figure 13.
Sum of prediction confidences for the scientific papers obtained from the digital archive of papers from Leibniz Institute for Catalysis.
Figure 13.
Sum of prediction confidences for the scientific papers obtained from the digital archive of papers from Leibniz Institute for Catalysis.
Figure 14.
Sum of margins between top two confidences for the scientific papers obtained from the digital archive of papers from Leibniz Institute for Catalysis.
Figure 14.
Sum of margins between top two confidences for the scientific papers obtained from the digital archive of papers from Leibniz Institute for Catalysis.
Table 1.
Types and counts of tags in their OWL files.
Table 1.
Types and counts of tags in their OWL files.
| Ontology Name | XML Classes | Number of Classes |
|---|
| AFO | rdfs:comment rdfs:label Literal | 2773 |
| NCIT | rdfs:comment rdfs:label | 1169 |
| SBO | rdfs:comment rdfs:label Literal | 534 |
| CHEBI | obo:IAO_0000115 rdfs:label | 35,067 |
| CHMO | rdfs:comment obo:IAO_0000115 rdfs:label | 2521 |
Table 2.
Sentence pairs of a new sentence from the scientific papers and the closest annotation from all the considered ontologies. The “carbon dioxide” annotation was assigned by BERT to all three above example new sentences. While BERT embeddings are more discriminative for the ontology classification task, the assigned sentences and low-dimensional embeddings on
Figure 5 indicate that this approach is more sensitive to the distribution shift problem.
Table 2.
Sentence pairs of a new sentence from the scientific papers and the closest annotation from all the considered ontologies. The “carbon dioxide” annotation was assigned by BERT to all three above example new sentences. While BERT embeddings are more discriminative for the ontology classification task, the assigned sentences and low-dimensional embeddings on
Figure 5 indicate that this approach is more sensitive to the distribution shift problem.
| | Sample 1 | Sample 2 | Sample 3 |
|---|
| New sentence | Moreover, there is an upper limit of operation, above which thermal decomposition will occur. | The difference is the main adsorption species during the reaction. | This enhancement of the Ni dispersion is very relevant because as reported in the literature NiO sites [...] |
| fastText closest | An end event specification is an event specification that is about the end of some process. | Reaction scheme where the products are created from the reactants [...] | The name of the individual working for the sponsor responsible for overseeing the activities of the study. |
| BERT closest | Carbon dioxide gas is a gas that is composed of carbon dioxide molecules. | Carbon dioxide gas is a gas that is composed of carbon dioxide molecules. | Carbon dioxide gas is a gas that is composed of carbon dioxide molecules. |
Table 3.
Zero-shot statistics for the distances of sentences to the closest ontology annotations.
Table 3.
Zero-shot statistics for the distances of sentences to the closest ontology annotations.
| Embeddings | Closest Distance Mean | Closest Distance Standard Deviation |
|---|
| fastText | 0.846 | 0.086 |
| BERT | 0.605 | 0.038 |
Table 4.
Hyperparameters of individual classifiers that were determined through grid-search on combinations of considered values. The bold values are selected values using a random stratified 5-fold cross-validation applied to a grid-search with the considered values.
Table 4.
Hyperparameters of individual classifiers that were determined through grid-search on combinations of considered values. The bold values are selected values using a random stratified 5-fold cross-validation applied to a grid-search with the considered values.
| Classifier | Hyperparameter | Considered and Selected Values |
|---|
| Random forest | bootstrap samples | {false, true} |
| criterion | {entropy, gini} |
| count of estimators | {5, 10, 15, 20, 25, 30} |
| fraction of features used in each split | {0.5, 0.7} |
| maximal depth | {5, 7, 9, 11} |
| Support vector machine | kernel type | {linear, radial basic} |
| kernel coefficient gamma | [0.001, 0.0001] |
| slack trade-off constant (C) | {1, 10, 100, 1000} |
| Gaussian process | kernel | {radial basic, dot product, matern, rational quadratic, white kernel} |
| K nearest neighbors | algorithm | {auto, ball tree, kd tree, brute} |
| distance metric exponent | {1, 2, 3, 4, 5} |
| number of considered neighbors | {1, 5, 9, 13, 17} |
| weights | {distance, uniform} |
| Multi-layer perceptron | activation function | {identity, logistic, relu, tanh} |
| hidden layer size | {1, 4, 16, 64} |
| learning rate for weights update | {adaptive, constant} |
| optimizer | {adam, lbfgs, sgd} |
| strength of L2 regularization term | {0.0001, 0.05} |
| random state | {0, 1} |
Table 5.
Statistic quality measures of the used classifiers with regards to the 20 testing datasets (mean [%] ± standard deviation [%]), where , , and (using the abbreviations T as True, F as False, P as Positive and N as Negative).
Table 5.
Statistic quality measures of the used classifiers with regards to the 20 testing datasets (mean [%] ± standard deviation [%]), where , , and (using the abbreviations T as True, F as False, P as Positive and N as Negative).
| | Accuracy | F1 | Precision | Recall |
|---|
| Gaussian process | | | | |
| K-nearest neighbor | | | | |
| Multi-layer perceptron | | | | |
| Random forest | | | | |
| Support vector machine | | | | |
Table 6.
Comparison of accuracy results on the 20 testing sets with ontology annotations. Counts of datasets are listed, where the model in the row has higher accuracy than the model in the column. The italic value marks counts, where the difference is not significant in the Wilcoxon test. The bold value marks a higher count from the pair, where the difference is significant.
Table 6.
Comparison of accuracy results on the 20 testing sets with ontology annotations. Counts of datasets are listed, where the model in the row has higher accuracy than the model in the column. The italic value marks counts, where the difference is not significant in the Wilcoxon test. The bold value marks a higher count from the pair, where the difference is significant.
| | Random Forest | Support Vector Machine | Gaussian Process | K-Nearest Neighbors | Multi-Layer Perceptron | Summary Score |
|---|
| RF | - | 0 | 0 | 0 | 0 | 0 |
| SVM | 20 | - | 2 | 15 | 13 | 50 |
| GP | 20 | 16 | - | 17 | 19 | 72 |
| k-NN | 20 | 3 | 3 | - | 5 | 31 |
| MLP | 20 | 4 | 1 | 14 | - | 39 |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).