Characterization of AMBN I and II Isoforms and Study of Their Ca2+-Binding Properties
Abstract
:1. Introduction
2. Results
2.1. AMBN ISO I and AMBN ISO II Belong to IDPs
2.2. AMBN ISO I and AMBN ISO II form Organized Oligomers
2.3. AMBN Oligomers Partially Disintegrate by Increasing Temperature
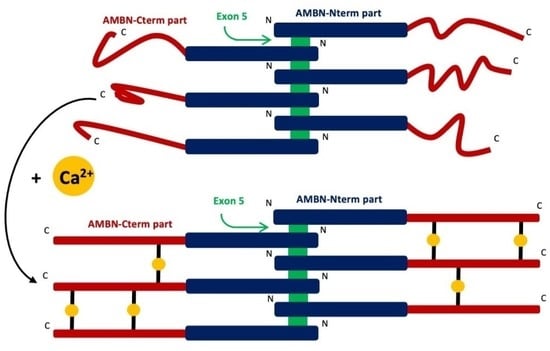
2.4. AMBN Binds Ca2+ Only in Its Oligomeric Form
3. Discussion
4. Materials and Methods
4.1. Design of AMBN Plasmid Constructs
4.2. Protein Expression and Purification
4.3. Circular Dichroism Spectroscopy
4.4. Dynamic Light Scattering
4.5. Analytical Ultracentrifugation
4.6. Differential Scanning Fluorimetry
4.7. Transmission Electron Microscopy
4.8. Capillary Electrophoresis
4.9. Microscale Thermophoresis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Ambn | Ameloblastin protein |
| AMBN | Human ameloblastin protein |
| AMBN del E5 | AMBN form lacking the exon 5 |
| AMBN ISO I | Isoform I of AMBN |
| AMBN ISO II | Isoform II of AMBN |
| AMBN-Cterm | C-terminal part of AMBN |
| AMBN-Nterm | N-terminal part of AMBN |
| Amel | Amelogenin |
| AUC | Analytical ultracentrifugation |
| BSA | Bovine serum albumin |
| BGE | Buffered background electrolyte |
| Ca2+ | Calcium ions |
| CD | Circular dichroism spectroscopy |
| CE | Capillary electrophoresis |
| D2P2 | Database of Disordered Protein Predictions |
| DLS | Dynamic light scattering |
| DMSO | Dimethylsulphoxide |
| DSF | Differential scanning fluorimetry |
| EGTA | Ethyleneglycol-bis(beta-aminoethylether)-N,N’-tetraacetic acid |
| EMPs | Extracellular matrix proteins |
| Exon 5 | Sequence encoded by exon 5 of AMBN |
| IDPs | Intrinsically disordered proteins |
| LB | Luria-Bertani broth |
| MMP20 | Enamelysin |
| PDI | Polydispersity index |
| Rh | Hydrodynamic radii |
| RS | Stokes hydrodynamic radii |
| SCPPs | Secretory Calcium-Binding Phosphoproteins |
| SEM | Standard errors of the mean |
| TEM | Transmission electron microscopy |
| TEV | Tobacco Etch Virus |
| Trx | Thioredoxin tag |
References
- Wald, T.; Bednarova, L.; Osicka, R.; Pachl, P.; Sulc, M.; Lyngstadaas, S.P.; Slaby, I.; Vondrasek, J. Biophysical characterization of recombinant human ameloblastin. Eur. J. Oral Sci. 2011, 119, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Wald, T.; Osickova, A.; Sulc, M.; Benada, O.; Semeradtova, A.; Rezabkova, L.; Veverka, V.; Bednarova, L.; Maly, J.; Macek, P.; et al. Intrinsically Disordered Enamel Matrix Protein Ameloblastin Forms Ribbon-like Supramolecular Structures via an N-terminal Segment Encoded by Exon 5. J. Biol. Chem. 2013, 288, 22333–22345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stakkestad, O.; Lyngstadaas, S.P.; Thiede, B.; Vondrasek, J.; Skalhegg, B.S.; Reseland, J.E. Phosphorylation Modulates Ameloblastin Self-assembly and Ca2+ Binding. Front. Physiol. 2017, 8, 10. [Google Scholar] [CrossRef]
- Boskey, A.L.; Villarreal-Ramirez, E. Intrinsically disordered proteins and biomineralization. Matrix Biol. 2016, 52–54, 43–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalmar, L.; Homola, D.; Varga, G.; Tompa, P. Structural disorder in proteins brings order to crystal growth in biomineralization. Bone 2012, 51, 528–534. [Google Scholar] [CrossRef]
- Grzybowska, E.A. Calcium-Binding Proteins with Disordered Structure and Their Role in Secretion, Storage, and Cellular Signaling. Biomolecules 2018, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Paine, M.L.; Snead, M.L. Protein Interactions During Assembly of the Enamel Organic Extracellular Matrix. J. Bone Min. Res. 1997, 12, 221–227. [Google Scholar] [CrossRef]
- Ravindranath, H.H.; Chen, L.-S.; Zeichner-David, M.; Ishima, R.; Ravindranath, R.M.H. Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem. Biophys. Res. Commun. 2004, 323, 1075–1083. [Google Scholar] [CrossRef]
- Deutsch, D.; Haze-Filderman, A.; Blumenfeld, A.; Dafni, L.; Leiser, Y.; Shay, B.; Gruenbaum-Cohen, Y.; Rosenfeld, E.; Fermon, E.; Zimmermann, B. Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: Brain and cells of the hematopoietic system. Eur. J. Oral Sci. 2006, 114, 183–189. [Google Scholar] [CrossRef]
- Deutsch, D.; Leiser, Y.; Shay, B.; Fermon, E.; Taylor, A.; Rosenfeld, E.; Dafni, L.; Charuvi, K.; Cohen, Y.; Haze, A. The human tuftelin gene and the expression of tuftelin in mineralizing and nonmineralizing tissues. Connect. Tissue Res. 2002, 43, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.C.-C.; Hu, Y.; Lu, Y.; Smith, C.E.; Lertlam, R.; Wright, J.T.; Suggs, C.; McKee, M.D.; Beniash, E.; Kabir, M.E. Enamelin is critical for ameloblast integrity and enamel ultrastructure formation. PloS ONE 2014, 9, e89303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Papagerakis, P.; Yamakoshi, Y.; Hu, J.; Bartlett, J.; Simmer, J. Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 2008, 389, 695–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruff, K.M.; Roberts, S.; Chilkoti, A.; Pappu, R.V. Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J. Mol. Biol. 2018, 430, 4619–4635. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Raduly, Z.; Miskei, M.; Fuxreiter, M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015, 589, 2533–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacDougall, M.; Simmons, D.; Gu, T.T.; Forsman-Semb, K.; Kärrman Mårdh, C.; Mesbah, M.; Forest, N.; Krebsbach, P.H.; Yamada, Y.; Berdal, A. Cloning, characterization and immunolocalization of human ameloblastin. Eur. J. Oral Sci. 2000, 108, 303–310. [Google Scholar] [CrossRef] [Green Version]
- Vymětal, J.; Slabý, I.; Spahr, A.; Vondrášek, J.; Lyngstadaas, S.P. Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur. J. Oral Sci. 2008, 116, 124–134. [Google Scholar] [CrossRef]
- Kärrman Mårdh, C.; Bäckman, B.; Simmons, D.; Golovleva, I.; Gu, T.T.; Holmgren, G.; MacDougall, M.; Forsman-Semb, K. Human ameloblastin gene: Genomic organization and mutation analysis in amelogenesis imperfecta patients. Eur. J. Oral Sci. 2001, 109, 8–13. [Google Scholar] [CrossRef]
- Putnam, C. PROTEIN CALCULATOR v3.4; The Scripps Research Institute: La Jolla, CA, USA, 2013. [Google Scholar]
- Simmer, J.P.; Fincham, A.G. Molecular Mechanisms of Dental Enamel Formation. Crit. Rev. Oral Biol. Med. 1995, 6, 84–108. [Google Scholar] [CrossRef] [Green Version]
- Margolis, H.C.; Beniash, E.; Fowler, C.E. Role of Macromolecular Assembly of Enamel Matrix Proteins in Enamel Formation. J. Dent. Res. 2006, 85, 775–793. [Google Scholar] [CrossRef]
- Paine, M.L.; White, S.N.; Luo, W.; Fong, H.; Sarikaya, M.; Snead, M.L. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001, 20, 273–292. [Google Scholar] [CrossRef]
- Mazumder, P.; Prajapati, S.; Lokappa, S.B.; Gallon, V.; Moradian-Oldak, J. Analysis of co-assembly and co-localization of ameloblastin and amelogenin. Front. Physiol. 2014, 5, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatakeyama, J.; Fukumoto, S.; Nakamura, T.; Haruyama, N.; Suzuki, S.; Hatakeyama, Y.; Shum, L.; Gibson, C.W.; Yamada, Y.; Kulkarni, A.B. Synergistic Roles of Amelogenin and Ameloblastin. J. Dent. Res. 2009, 88, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Kegulian, N.C.; Bapat, R.A.; Moradian-Oldak, J. Ameloblastin Binds to Phospholipid Bilayers via a Helix-Forming Motif within the Sequence Encoded by Exon 5. ACS Omega 2019, 4, 4405–4416. [Google Scholar] [CrossRef] [PubMed]
- Wald, T.; Spoutil, F.; Osickova, A.; Prochazkova, M.; Benada, O.; Kasparek, P.; Bumba, L.; Klein, O.D.; Sedlacek, R.; Sebo, P.; et al. Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc. Natl. Acad. Sci. USA 2017, 114, E1641–E1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.; Chandrababu, K.B.; Moradian-Oldak, J. Ameloblastin peptide encoded by exon 5 interacts with amelogenin N-terminus. Biochem. Biophys. Rep. 2016, 7, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Li, M.; Xu, X.; Xiong, J.; Huang, C.; Zhang, X.; Hu, A.; Peng, L.; Cai, D.; Zhang, L.; et al. Whole exome sequencing identifies an AMBN missense mutation causing severe autosomal-dominant amelogenesis imperfecta and dentin disorders. Int. J. Oral Sci. 2018, 10, 26. [Google Scholar] [CrossRef]
- Delgado, S.; Davit-Béal, T.; Allizard, F.; Sire, J.-Y. Tooth development in a scincid lizard, Chalcides viridanus (Squamata), with particular attention to enamel formation. Cell Tissue Res. 2005, 319, 71–89. [Google Scholar] [CrossRef]
- Delgado, S.; Casane, D.; Bonnaud, L.; Laurin, M.; Sire, J.-Y.; Girondot, M. Molecular Evidence for Precambrian Origin of Amelogenin, the Major Protein of Vertebrate Enamel. Mol. Biol. Evol. 2001, 18, 2146–2153. [Google Scholar] [CrossRef] [Green Version]
- Girondot, M.; Sire, J.-Y. Evolution of the amelogenin gene in toothed and toothless vertebrates. Eur. J. Oral Sci. 1998, 106, 501–508. [Google Scholar] [CrossRef]
- Fernàndez-Busquets, X.; Körnig, A.; Bucior, I.; Burger, M.M.; Anselmetti, D. Self-Recognition and Ca2+-Dependent Carbohydrate–Carbohydrate Cell Adhesion Provide Clues to the Cambrian Explosion. Mol. Biol. Evol. 2009, 26, 2551–2561. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.S. “Liquid-like” biomineralization protein assemblies: A key to the regulation of non-classical nucleation. CrystEngComm 2013, 15, 8388–8394. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yamakoshi, Y.; Hu, J.C.-C.; Gomi, K.; Arai, T.; Fukae, M.; Krebsbach, P.H.; Simmer, J.P. Splicing Determines the Glycosylation State of Ameloblastin. J. Dent. Res. 2007, 86, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Yamakoshi, Y.; Richardson, A.S.; Nunez, S.M.; Yamakoshi, F.; Milkovich, R.N.; Hu, J.C.C.; Bartlett, J.D.; Simmer, J.P. Enamel proteins and proteases in Mmp20 and Klk4 null and double-null mice. Eur. J. Oral Sci. 2011, 119, 206–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztányi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D2P2: Database of disordered protein predictions. Nucleic Acids Res. 2012, 41, D508–D516. [Google Scholar] [CrossRef] [Green Version]
- Kjaergaard, M.; Nørholm, A.-B.; Hendus-Altenburger, R.; Pedersen, S.F.; Poulsen, F.M.; Kragelund, B.B. Temperature-dependent structural changes in intrinsically disordered proteins: Formation of alpha-helices or loss of polyproline II? Protein Sci. 2010, 19, 1555–1564. [Google Scholar] [CrossRef]
- Erickson, H.P. Size and Shape of Protein Molecules at the Nanometer Level Determined by Sedimentation, Gel Filtration, and Electron Microscopy. Biol. Proced. Online 2009, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- Heegaard, N.H.H.; Robey, F.A. A capillary electrophoresis-based assay for the binding of Ca2+ and phosphorylcholine to human C-reactive protein. J. Immunol. Methods 1993, 166, 103–110. [Google Scholar] [CrossRef]
- Clapham, D.E. Calcium Signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [Green Version]
- Ikura, M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 1996, 21, 14–17. [Google Scholar] [CrossRef]
- Sheng, Z.-H.; Rettig, J.; Cook, T.; Catterall, W.A. Calcium-dependent interaction of N-type calcium channels with the synaptic core complex. Nature 1996, 379, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Christopeit, T.; Gossas, T.; Danielson, U.H. Characterization of Ca2+ and phosphocholine interactions with C-reactive protein using a surface plasmon resonance biosensor. Anal. Biochem. 2009, 391, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Beyeler, M.; Schild, C.; Lutz, R.; Chiquet, M.; Trueb, B. Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp. Cell Res. 2010, 316, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, R.M.; Devarajan, A.; Uchida, T. Spatiotemporal expression of ameloblastin isoforms during murine tooth development. J. Biol. Chem. 2007, 282, 36370–36376. [Google Scholar] [CrossRef] [Green Version]
- Stakkestad, Ø.; Lyngstadaas, S.P.; Vondrasek, J.; Gordeladze, J.O.; Reseland, J.E. Ameloblastin peptides modulates the osteogenic capacity of human mesenchymal stem cells. Front. Physiol. 2017, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, G.; Varadarajan, R. Facile measurement of protein stability and folding kinetics using a nano differential scanning fluorimeter. Protein Sci. 2019, 28, 1127–1134. [Google Scholar] [CrossRef]
- Rozbeský, D.; Kavan, D.; Chmelík, J.; Novák, P.; Vaněk, O.; Bezouška, K. High-level expression of soluble form of mouse natural killer cell receptor NKR-P1C (B6) in Escherichia coli. Protein Expr. Purif. 2011, 77, 178–184. [Google Scholar] [CrossRef]
- Hayes, D.; Laue, T.; Philo, J. Program Sednterp: Sedimentation Interpretation Program; Alliance Protein Laboratories: Thousand Oaks, CA, USA, 1995. [Google Scholar]
- Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000, 78, 1606–1619. [Google Scholar] [CrossRef] [Green Version]
- Brautigam, C.A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 562, pp. 109–133. [Google Scholar]
- Štěpánová, S.; Václav, K. Capillary Electrophoretic Methods Applied to the Investigation of Peptide Complexes. J. Sep. Sci. 2015, 38, 2708–2721. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]




| Protein | 0 mM CaCl2 | 10 mM CaCl2 | ||
|---|---|---|---|---|
| Rh (nm) | PDI | Rh (nm) | PDI | |
| AMBN ISO I | 24.0 | 0.13 | 21.6 | 0.09 |
| (0.2) | (0.02) | (0.2) | (0.08) | |
| AMBN ISO II | 17.7) | 0.12 | 16.9 | 0.09 |
| (0.1) | (0.01) | (0.1) | (0.01) | |
| AMBN del E5 | 5.7 | 0.06 | 5.7 | 0.06 |
| (0.05) | (0.01) | (0.05) | (0.01) | |
| BSA | 5.03 | 0.30 | NA | NA |
| (0.03) | (0.01) | |||
| Protein | 0 mM CaCl2 | 10 mM CaCl2 | ||||
|---|---|---|---|---|---|---|
| RS (nm) | s20,w (S) | f/f0 | RS (nm) | s20,w (S) | f/f0 | |
| AMBN ISO I monomer | 4.44 | 2.53 | 2.09 | 4.30 | 2.61 | 2.02 |
| AMBN ISO I oligomers | 15.5 | 26.1 | 1.70 | 15.4 | 28.5 | 1.64 |
| AMBN ISO II monomer | 4.28 | 2.53 | 2.05 | 4.10 | 2.65 | 2.18 |
| AMBN ISO II oligomers | 13.5 | 19.2 | 1.72 | 13.0 | 20.9 | 1.63 |
| AMBN del E5 | 4.98 | 1.98 | 2.20 | 4.85 | 2.03 | 2.10 |
| BSA | 3.38 | 4.60 | 1.27 | NA | NA | NA |
| Protein | Kb ± SD a (L/mol) | mp,eff,0 ± SD a (10−9 m2 V−1 s−1) | mPM ± SD a (10−9 m2 V−1 s−1) | R2 | KD ± SD a (mol/L) |
|---|---|---|---|---|---|
| AMBN ISO I | 56.8 ± 6.3 | −13.8 ± 0.2 | −2.39 ± 0.51 | 0.993 | 0.017 ± 0.002 |
| AMBN ISO II | 54.4 ± 3.0 | −14.2 ± 0.2 | −2.36 ± 0.27 | 0.998 | 0.018 ± 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vetyskova, V.; Zouharova, M.; Bednarova, L.; Vaněk, O.; Sázelová, P.; Kašička, V.; Vymetal, J.; Srp, J.; Rumlová, M.; Charnavets, T.; et al. Characterization of AMBN I and II Isoforms and Study of Their Ca2+-Binding Properties. Int. J. Mol. Sci. 2020, 21, 9293. https://doi.org/10.3390/ijms21239293
Vetyskova V, Zouharova M, Bednarova L, Vaněk O, Sázelová P, Kašička V, Vymetal J, Srp J, Rumlová M, Charnavets T, et al. Characterization of AMBN I and II Isoforms and Study of Their Ca2+-Binding Properties. International Journal of Molecular Sciences. 2020; 21(23):9293. https://doi.org/10.3390/ijms21239293
Chicago/Turabian StyleVetyskova, Veronika, Monika Zouharova, Lucie Bednarova, Ondřej Vaněk, Petra Sázelová, Václav Kašička, Jiri Vymetal, Jaroslav Srp, Michaela Rumlová, Tatsiana Charnavets, and et al. 2020. "Characterization of AMBN I and II Isoforms and Study of Their Ca2+-Binding Properties" International Journal of Molecular Sciences 21, no. 23: 9293. https://doi.org/10.3390/ijms21239293