Abstract
In freshwater systems, cyanobacteria are strong competitors under enhanced temperature and eutrophic conditions. Understanding their adaptive and evolutionary potential to multiple environmental states allows us to accurately predict their response to future conditions. To better understand if the combined impacts of temperature and nutrient limitation could suppress the cyanobacterial blooms, a single strain of Microcystis aeruginosa was inoculated into natural phytoplankton communities with different nutrient conditions: oligotrophic, eutrophic and eutrophic with the addition of bentophos. We found that the use of the bentophos treatment causes significant differences in prokaryotic and eukaryotic communities. This resulted in reduced biodiversity among the eukaryotes and a decline in cyanobacterial abundance suggesting phosphorus limitation had a strong impact on the community structure. The low temperature during the experiment lead to the disappearance of M. aeruginosa in all treatments and gave other phytoplankton groups a competitive advantage leading to the dominance of the eukaryotic families that have diverse morphologies and nutritional modes. These results show cyanobacteria have a reduced competitive advantage under certain temperature and nutrient limiting conditions and therefore, controlling phosphorus concentrations could be a possible mitigation strategy for managing harmful cyanobacterial blooms in a future warmer climate.
Similar content being viewed by others
Introduction
Freshwater environments support nearly 6% of all described species yet only make up 0.01% of the planet’s water and cover approximately 0.8% of the Earth’s surface 1. Freshwater lakes provide a rich biodiversity making them an invaluable natural resources in ecological, economic, scientific and cultural terms2. However, a rapid expansion of the world’s population and associated increases in industrialisation, land use changes and environmental degradation, has had a detrimental impact on freshwater lakes3. Nutrient enrichment, specially of nitrogen (N) and phosphorus (P), both promote eutrophication and cyanobacterial algal blooms (CyanoHABS; one of the few groups able to dominate the entire primary producer community) in freshwater lakes4,5. Coincidently, the rapid eutrophication also corresponds with freshwater lakes coming under pressure from climatic drivers, such as increased warming and changes in the hydrological cycle6. Over the last century, global warming has become one of the most serious issues facing freshwater environments, with temperatures in temperate European lakes increasing on average by 0.58 °C decade−17. Global warming and eutrophication are considered two of the most important causes to the occurrence of cyanobacteria blooms in freshwater aquatic ecosystems8,9.
Eutrophication caused by human activities can cause significant shifts in freshwater primary producer communities towards fast growing species such as cyanobacteria resulting in harmful algal blooms10,11. Concordantly, unprecedented rates of global warming in recent decades has also led to a rise in the frequency of CyanoHABS in temperate lakes8,12 as they exhibit optimal growth rates at temperatures greater than 25 °C13 and compete most effectively against eukaryotic algae, resulting in a decline in diversity9. Increased temperatures can also exacerbate the problem of bottom water anoxia and internal loading of nutrients especially P, enhancing stratification, which can further fuel CyanoHABs14. Therefore, reducing P is an important factor in controlling eutrophication and mitigating algal blooms such as CyanoHABs15.One cost-effective approach is the use of bentophos (also known as PhosLock), a patented lanthanum-modified bentonite clay formation which binds with bioavailable P, removing it from the system16. Previous studies have shown the ability of bentophos to reduce P in the water column, as an effective measure to manage eutrophication17 and to reduce CyanoHABs18. Furthermore, Bishop and Richardson19 found the use of bentophos caused a shift in cyanobacteria abundance, composition and density whilst Lang et al.17 found a relative increase in the proportion of non-cyanobacteria planktonic algae such as cryptophytes, chlorophytes and diatoms. However, there is limited work on how the use of bentophos influences freshwater communities at a genus and species level. Therefore, further research is needed into how it affects freshwater ecosystem and whether it can ‘revert’ them back to a pre-eutrophication state.
A well-known bloom forming cyanobacterial genus is Microcystis, a strong competitor against other phytoplankton groups such as chlorophytes, cryptophytes and siliceous algae under elevated nutrients and especially temperature conditions20. Generally, it is predicted that cyanobacteria will increase in dominance under climate change13, though empirical data on their evolutionary responses are scarce. Although, it appears that cyanobacteria vary in their response to higher temperatures over a wide range of nutrient levels and light intensities13,21. Preliminarily mesocosm results suggest that cyanobacteria (specially Microcystis) have the strong potential to cope under warming conditions and that previous exposure to warming was a critical factor in the phytoplankton community composition22. The use of mesocosm experiments allows for the assessment of the competitive ability of ‘adapted’ species within natural communities over multiple trophic states23, especially as recent studies suggest that cyanobacteria may have a reduced competitive advantage under nutrient limiting conditions24. The use of a common cyanobacterial species (such as Microcystis aeruginosa) adapted to future conditions could help determine how cyanobacteria and other algae groups will react to the use of P remediation strategies such as bentophos. Utilising mesocosms with different nutrient conditions, allows us to directly compare the effects on microbial and algal communities and to test if bentophos can improve the water quality of eutrophic freshwater systems.
This research aims to assess the combined effects of temperature and nutrient limitation on cyanobacterial populations. Firstly, we tested the impact of nutrient limitation using three different environments, (1) oligotrophic, (2) eutrophic and (3) eutrophic with the addition of bentophos on microbial and algal communities to determine if it gave cyanobacteria a competitive advantage against other algae groups. Secondly, we analysed the impact of temperature by using a M. aeruginosa strain adapted to ambient (22 °C, mean summer temperature) and a heated temperature (26 °C, the predicted mean temperature by 2100) on natural planktonic communities to determine if further adaptation gives certain cyanobacterial species such as M. aeruginosa a competitive advantage. Thirdly, we then compared these results with the mesocosm experiments carried out in 201922 with the same strain of M. aeruginosa, to determine if the concordant effects of nutrient limitation and temperature result in different phytoplankton communities.
Results
Growth rate evolution
The growth rates of M. aeruginosa strain (M11) grown for 2 years at 22 °C (ambient) and 26 °C (heated) were measured in the control environment (22 °C) every 8 months (Months 0, 8, 16 and 24). This was to determine if adaptation resulted in enhanced growth under ambient temperature over time. At Month 0, the M11 growth rates were similar (μ = 0.156–0.159 day−1; Fig. 1A). The growth rates for the strain grown at both temperatures fluctuated, showing no pattern after 2 years of exposure, with similar growth rates to those measured at the beginning of the experiment (Month 0). At the end of the experiment (Month 24) the growth rate of the ambient and heat-adapted M11 showed no significant differences in growth rate (as both were measured at 22 °C). The differences between growth rates between the ambient and heat-adapted M11 was statistically significant only for Month 8 (p < 0.05).
(A) Mean Growth rate and standard deviation of M. aeruginosa (strain M11) measured every 8 months for ambient (M11_22) and heat-adapted (M11_26) and (B) the thermal reaction norms with standard deviation showing the growth rate at 2 °C intervals between 20 and 40 °C for both ambient and heat-adapted M11.
The TRNs results showed clear differences between the growth rates over a range of temperatures for both ambient and heat-adapted M11 (Fig. 1B). The lethal temperature was 2 °C higher for heat-adapted compared to ambient-adapted M11. The growth rates for ambient-adapted M11 were stable until 32 °C, declining until 37 °C before reaching lethal temperature at 38 °C. The growth rates for heat-adapted M11 were constant until 39 °C before reaching lethal temperature at 40 °C.
qPCR gene expression
Candidate genes were selected from different pathways involved in heat shock response or thermal tolerance in cyanobacteria25. The qPCR analysis results were calculated as fold change in the expressions of target genes (2−ΔΔCt): cya1, pnp, pyrR, clpC1, clpC2, and sigF within the specific time period and samples (Months 0, 8, 16 and 24 for ambient- and heat adapted M11) relative to the M. aeruginosa M11 rnpB (reference gene). Ambient adapted M11 showed a reduction in gene expression levels with 0.3–1.9-fold reduction by Month 24 (Fig. 2). The expression of clpC1showed an up-regulation trend in comparison to all other genes of interest. Heat adapted M11 displayed an overall upregulation over the time period of the experiment, with an approximately 1.8–5.6-fold increase in gene expression of clpC1. By Month 24, the largest increase was observed for clpC1 gene with a > fivefold gene expression within heat adapted M11, demonstrating a steady increase within this study (Fig. 2). The cya1 gene profiles showed the least change of expression for both ambient and heat adapted M11, with a slight down-regulation.
The gene expression results for six genes for M11_22 and M11_26 for Months 0–24. The values represent the relative abundance of target genes based on rnpB, which was used as reference gene. These results were normalized against Month 0_M11_22 to determine if temperature has caused a change in gene expression. Each test was done in three replicates.
Land-based mesocosm experiments - temperature and light intensity
The fluctuations in temperature of the three treatments were similar throughout the whole mesocosm experiment. The mean temperature throughout the first 14 days of the experiment averaged c.21–22 °C before declining in all treatments in the third week to 16 °C (Fig. S1A). In the fourth week, the water temperature increased back to an average of c.21–22 °C. The mesocosms with the oligotrophic treatment consistently had a daytime temperature approx. 1 °C higher than the other two treatments. Oligotrophic systems have lower algae abundance and higher light intensity at depth (reflected in the light intensity measurements taken during this study) due to greater water transparency, causing a slightly elevated temperature.
The light intensity measurements were highest in the oligotrophic compared to the other two treatments (Fig. S1B). The light intensity measurements were relatively stable throughout the four-week experiment but there was a slight dip in week 3 (coinciding with the decline in temperature) for all treatments.
Nutrients
Concentrations for all nutrients were lowest in the oligotrophic treatment (Table S1). The eutrophic treatment had the highest levels of phosphorus (4.20 μgL−1 for PO4-P; 35.41 μgL−1 for TP) while the use of bentophos led to a reduction in P (to 2.87 μgL−1 for PO4-P; 19.18 μgL−1 for TP). The bentophos treatment had the highest concentration of both NO2 (16.55 μgL−1) and NO3− (732.01 μgL−1).
Phytoplankton group dynamics
Chlorophyll a concentration (µL-1) increased as the experiment progressed, with the largest increase seen in the eutrophic treatment and the smallest in the bentophos treatment (Fig. 3). There were significant differences (using ANOVA) between Week 1 and Week 4 concentrations (p < 0.001, = 16.637; Table S2). The greatest increase was observed between Week 1 and Week 2 (p = 0.014, F = 16.637), with a smaller increase but still a significant rise through Weeks 2–4 (p = 0.002, F = 16.637). The eutrophic treatment showed the largest increase in mean chlorophyll a concentrations of 9.04 (Week 1) to 23.34 μgL−1 (Week 4). While the oligotrophic and bentophos treatments saw an increase in mean chlorophyll a of 4.02–9.10 μgL−1 and 8.67–11.85 μgL−1 respectively over the same time period. No significant differences or increases were observed in chlorophyll a concentration between the control and the mesocosms inoculated with M11_22 and M11_26 (Fig. 3).
The most noteworthy result was that there were no discernible differences between the brown group, cyanobacteria and cryptophyte concentrations throughout the four-week experiment, with chlorophytes showing the only significant change in concentrations (p < 0.001). Mean chlorophyte concentration increased significantly in all treatments as the experiment progresses, with the largest increase in the eutrophic mesocosm (6.86–20.48 μgL−1; Fig. 3). Mean chlorophyte concentrations increased 4.76 and 3.33 μgL−1 for the oligotrophic and bentophos treatments respectively. In general, the groups of microalgae were not affected by the inoculation of ambient or heat-adapted M. aeruginosa, with similar composition among all three treatments. The mean cyanobacteria concentration was slightly higher in the eutrophic treatment compared to the other two treatments, with the highest concentration in the mesocosms inoculated with heat-adapted M11, followed by ambient-adapted M11 and then the control (Fig. 3C).
16S and 18S metabarcoding data
A total of 1.0 million sequence reads were obtained following the 16S rDNA amplicon sequencing and 1.4 million for the 18S rDNA. A number of 503,557 reads (248,090 for 16S and 255,467 for 18S) passed the processing and filtering steps (sequencing quality and read length). Following reassembly, alignment clean-up and mapping, the final abundance of OTUs from the nine samples contained 520 prokaryotic OTUs and 371 eukaryotic OTUs.
The three treatments had a similar composition of bacterial communities, yet the abundances differed (Fig. 4A; Table S3). The most abundant prokaryotic OTUs identified belong to the phylum Proteobacteria (28.2–37.0%), with a majority of these OTUs assigned to Alphaproteobacteria (22.7–30.5%) and Gammaproteobacteria (3.6–5.6%) (Fig. 4A). The main difference between the three treatments was a lower abundance of Planctomycetes in the bentophos treatment (< 10.7%) compared to the oligotrophic (11.8–25.9%) and the eutrophic treatments (10.3–22.9%), with the majority consisting of the orders Pirellulales and Planctomycetales (Fig. S2). Higher concentrations of Actinobacteria (consisting of 90% Frankiales) were found in the eutrophic treatment inoculated with heat adapted M. aeruginosa (17.8%) compared to the samples inoculated with ambient M. aeruginosa and the oligotrophic tanks (< 5.8%). There was a high abundance of Bacteroidetes (15.9–23.5%) observed in all samples with no differences between treatments or the strain of M. aeruginosa. Verrucomicrobia was observed in higher abundances in the oligotrophic and eutrophic treatments (8.7–18.2%) compared to bentophos treatment (6.7–10.4%).
Cyanobacteria were found in low abundances in all samples (3.3–6.7%), with no differences between the samples inoculated with M. aeruginosa and the control samples (Fig. 4A). Even though the samples were inoculated with M. aeruginosa (except the control samples), no Microcystis was present in any of the samples. Most cyanobacterial DNA reads were assigned to the genus Pseudanabaena and Synechococcus (Fig. S2), however, due to sequence ambiguity, individual species could not be identified. Chloroflexi (a phylum of thermophilic bacteria) were found in slightly higher abundances in the tanks inoculated with heat adapted Microcystis (0.3–1.0%) compared to other tanks (< 0.2%). A higher proportion of the DNA sequences from the bentophos treatment (11.18–22.66%) compared to < 6.99% in the other treatments remained unassigned.
The eukaryotic populations in all samples were dominated by the SAR supergroup (a clade that includes Stramenopiles, Alveolates and Rhizaria;26) with abundances of 56.4–80.3% (Fig. 4B; Table S4). The majority of the SAR supergroup consists of the family Ochrophyta 42.0–68.4% of total abundance, followed by Ciliophora (2.3–18.4%) and Cercozoa (0.7–5.6%; Fig. S3). There was no discernable pattern between Ochrophyta, Ciliophora and Cercozoa abundance and a treatment/Microcystis inoculation. The second most dominant group was from the order Metazoa consisting mostly of the family Copepoda (0.7–24.9%) followed by Monogonota (0.6–3.1%). A high abundance of copepods was observed in samples Oligotrophic_M11_22, Oligotrophic_Contol, Eutrophic_Control, Bentophos_M11_22 and Bentophos_M11-26 (13.0–24.9%) compared to < 5.1% in the remaining samples (Fig. 4B). Chlorophytes were present in all samples with abundances ranging from 2.3 to 7.4%. Low abundances of fungi (mostly belonging to the order Cryptomycota) were identified in all samples with the highest abundance found in the bentophos treatment. Dinoflagellates were present in low abundances in all samples (< 3.2%). A variable proportion of reads (1.8–7.8%) were unassigned.
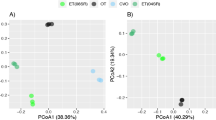
The PCoA of the bacterial communities showed a clear difference between the bentophos and the other two treatments (Fig. 5), which were distinguished by higher abundances (compared to the oligotrophic and eutrophic treatments) of bacteria only found in metagenomic data (Hydrogenedentes, Dependentiae, Gemmatimonadetes and Patescibacteria) and a lack of cyanobacterial taxa (Fig. S4). The remaining two groups consisted of a mixture of the oligotrophic and eutrophic treatments characterised by high abundances of Planctomycetes, cyanobacteria, BRC1 and Spirochaetes. The separation of bentophos treatment along both the first and second axis showed the impact of the bentophos on the bacterial communities’ composition (Fig. S4A). Compared to the 16S PCA, there were no clear groupings between treatment or the isolate of M11 used in the 18S PCA due to the dominance of Ocrophyta and Ciliophora (Fig. S4).
The variation in terms of beta diversity (using weighted Unifrac distances) was explained by the treatment specifically shown by the clear grouping of the bentophos treatment, as shown by axis 1 (which explained 59.10% of the variance). There were no other clear groupings for the other two treatments nor between the isolate of M11 used (Fig. 5).
Comparison with previous experimental mesocosm results
Both the 201922 and 2020 mesocosm experiments had a similar bacterial composition but different abundances. Compared to the 2019 experiment, the 2020 mesocosms displayed a lower abundance of proteobacteria, planctomycetes, and especially cyanobacteria. The latter had a much smaller abundance of 3.3–6.7% compared to 2019 (20–50%). The main difference between the two experiments is the lack of dominance of the SAR supergroup in the 2019 experiment, with the most abundant group being Metazoa consisting of copepods (7–30% of total reads) and gastrotrichs (7–33% of total reads).
Discussion
The incorporation of long-term adapted M. aeruginosa into short-term mesocosm experiments can help explore the mechanisms involved in the adaptation process27 and see if increased temperature gives the strain a competitive advantage over natural phytoplankton communities. Analysis of the qPCR gene expression of M. aeruginosa showed an increased upregulation of genes associated with enhanced thermal tolerance (Fig. 2). A continuous increase in upregulation in the heat-adapted M11 (but not the ambient-adapted M11) over time of the genes clpC1 and pnp suggests adaptation to increased temperature ‘improved’ over time. There was also an increase in the expression of sigF and pyrR genes although this was not proportional to the level of adaption. Tillich et al.25 found that the clpC1, pnp, sigF and pyrR genes gave the highest fitness advantage at enhanced temperatures and are important for increased thermal tolerance in Synechocystis. These results suggest that the enhanced expression and upregulation of these genes gave heat-adapted M11 an advantage over the ambient-adapted M11in warmer temperatures. This is supported by a former mesocosm experiment 22, who found that high temperatures (30 °C) gave heat-adapted M. aeruginosa a strong competitive advantage over other algal groups leading to significantly higher concentrations. However, we were not able to replicate these results as low temperatures for the duration of this experiment most likely lead to the disappearance of both ambient and heat adapted M. aeruginosa. Nevertheless, we found that the use of bentophos lead to a substantial reduction in cyanobacterial taxa suggesting that shifting P availability could help reduce the group’s competitive ability under warming conditions.
Interestingly, the bentophos treatment showed significant differences from the other two treatments suggesting P limitation has impacted algal and bacterial communities in freshwater lakes (Fig. 5). These samples were characterised by a lack of cyanobacterial taxa (3.3–4.0%) and higher abundances of small celled non-autotroph bacteria only identified in metagenomic data (Hydrogenedentes, Dependentiae, Gemmatimonadetes and Patescibacteria; Fig. S4A). Limited information on the ecology of these bacteria suggests they are generalists, as they have been found widespread across diverse environments28,29,30. Also, Patescibacteria have a PhoR-PhoB two component regulatory system, which is advantageous in low P environments31. Moreover, the eukaryotic communities did not show a clear difference between the bentophos and the other treatments, which were characterised by taxa which comprised of < 1% of the relative abundance (Goniomonas, choanoflagellida and Diphylleia rotans; Fig. S4B). These orders are cosmopolitan species found in a wide range of freshwater environments with varying nutrient regimes32,33,34. Multiple studies have found that the use of bentophos in temperate lakes led to a significant reduction in cyanobacteria and a shift away from cyanobacteria dominance17,35. bentophos seems to have a greater impact on cyanobacteria compared to eukaryotic planktonic groups, as N2 fixing cyanobacteria (which bloom under N limited conditions, common in eutrophic lakes36) have a stronger P requirement35,37. This could be a potential reason for the lack of N2 fixing cyanobacteria residence populations. It could be suggested that the use of bentophos (which binds to the available P preventing it from being utilised by microalgae and bacteria), altered the N:P ratio (to 38.2; Table S1) leading to P limitation. This could have led to a reduction in cyanobacteria taxa, a rise in generalist small cell bacteria and Patescibacteria) who have developed the ability to survive under low P conditions.
The cyanobacteria taxa in all samples (3.3–6.7%) were dominated by the genera Synechococcus and Pseudanabaena due to their ability to compete well under nutrient limiting and varying light intensities38. Studies from temperate freshwater lakes (Tiefer See, Germany38) found a dominance of Synechococcus in the late Spring/early Summer due to their physiological conditions such as rapid growth, ability to adapt to changing light conditions and affinity to low chlorophyll a conditions39. Under nutrient limiting conditions, Synechococcus has the ability to utilise orthophosphate and other organic phosphorous sources apart from inorganic phosphates and to store N40, enhancing its competitive ability against other phytoplankton groups41. Surprisingly the bentophos treatments (2.0–3.1%) did not have the highest relative abundance of Synechococcus, with this treatment being characterised by low abundance (Fig. S2). Even though Synechococcus has an extreme adaptability to a wide range of temperatures, it has improved growth at higher temperatures42,43 suggesting the low temperatures during the experiment dampened their growth. Synechococcus success in oligotrophic treatments could be due to its capacity to adapt to high light conditions in oligotrophic environments43. Some strains of Pseudanabaena spp. have the ability to regulate the ratio of the accessory photosynthetic pigments phycocyanin and phycoerythrin, in response to changing light wavelengths44 helping them to adapt to the prevailing light spectrum45. This could explain why the oligotrophic treatment with the highest light intensity had the highest abundance of Pseudanabaena spp (0.8–5%). Nevertheless, similar abundances in the eutrophic conditions suggest that there must be other contributing factors. Abiotic factors that could explain this include grazing by ciliates and protists, viral lysis and nutrient availability46,47.
Surprisingly, even though M. aeruginosa were inoculated into all mesocosms (except the controls), it had disappeared by the end of the four-week experiment (Fig. S2). Whereas, in the 2019 mesocosm experiment, M. aeruginosa (M11) consisted of 49% OTUs (in ambient tanks) and 36% OTUs in heated tanks22. M. aeruginosa is a strong competitor under increased temperatures but growth is suppressed when the temperature is < 20 °C48. Also, the analysis of temperature-dependent growth rates determined that many eukaryotic algae had maximised or declining growth rates at > 20 °C, whereas the growth rate of many cyanobacteria species increased13,49. Previous mesocosm, laboratory and larger-scale limnological studies have also found that Microcystis spp. yielded increased growth rates under enhanced temperatures50,51,52. This is further supported by the 2019 mesocosm experiment22, which had temperatures up to 32–33 °C, most likely aiding the dominance of M. aeruginosa13. This suggests that the low temperature (< 22 °C) recorded in the mesocosms could have reduced their competitive ability and limited its growth, resulting in the disappearance of M. aeruginosa53.
All treatments showed a similar bacterial community composition after the four-week mesocosm experiment (Fig. 4A; Table S5). Proteobacteria, a group of gram-negative bacteria, dominated bacterial populations in all treatments suggesting its relative abundance was not influenced by trophic state or nutrient limitation. Similar relative abundances of proteobacteria were expected as they have been observed in a wide range of freshwater environments (Fig. 4A;54). This could also explain why proteobacteria dominated in the 2019 mesocosm despite the different nutrient and temperature conditions22. Bacteroidetes was the second most abundant phylum (15.6–23.5%) but there were no significant differences between treatments (Fig. 4A) providing further evidence that this phylum has been found in a diverse range of freshwater habitats55,56. There were lower relative abundances of planctomycetes observed in the bentophos treatment (9.5–10.7%) compared to the oligotrophic (11.8–25.9%) and eutrophic (10.3–22.9%) treatments (Fig. 4A). Planctomycetes are particle-associated and mostly grow in nutrient poor oligotrophic environments but have also been found in eutrophic conditions57. They have a significant impact on lake biogeochemical cycles through their ability to mineralise and breakdown detritus particles in the water column, making nutrients available to other organisms29,57. The reduced availability of nutrients in the oligotrophic treatment (which are characterised by higher abundances of Planctomycetes; Fig. S4A) could also explain the lower abundances in the 2019 mesocosm experiment22 and why this treatment also had higher relative abundances of Verrucomicrobia55,56. Schmidt et al.56 found that in a collection of freshwater lakes this phylum was significantly over-represented in oligotrophic (relative to eutrophic) lakes. Studies have found that temperature and total phosphorus (TP) are the main factors influencing bacterial communities in freshwater lakes58. The low and consistent temperatures within all mesocosms (Fig. S1) and the lack of thermophilic prokaryotes such as chloroflexi59 suggests that nutrient concentrations including phosphorus are most likely driving the differences between microbial communities in this experiment. Low abundances of Firmicutes in all mesocosms (< 0.1%) could be due to aerobic conditions and low cyanobacterial abundances (Fig. 4A), as this family exhibits a significant advantage during the anaerobic decomposition of cyanobacteria blooms60. Other phyla occurred in low relative abundances (< 3.0%) probably due to a combination of physical, biological and chemical factors61.
The eukaryotic diversity (Fig. 4B; Table S5) in all samples was dominated by the protists from the SAR supergroup specially with the families Orchrophyla, Ciliophora and Cercozoa. The family Orchrophyla were dominated by the genera Chrysamoeba and Ochromonas whereas the families Ciliophora and Cercozoa had more diverse genera present (Fig. S3). The genera (Chrysamoeba and Ochromonas) and the family Ciliophora, encompasses a broad range of morphological diversity and a variety of trophic modes within the microbial food web62. A likely reason for their dominance is that the species within both genera can have different nutritional modes63,64. Generally, mixotrophy enables organisms to overcome unfavourable conditions allowing them to inhibit environments with altered N:P ratios and high light intensities65. Mixotrophs have an advantage over heterotrophs under high light intensities, as they can acquire nutrients via phototrophy66. Also, mixotrophs have higher cellular N contents (compared to heterotrophs) and therefore a higher demand for N, so are more successful under P limiting conditions67. This could explain why there is similar relative abundance of these genera across all treatments regardless of nutrient concentrations or light intensity. Cercozoa is a heterotrophic predatory flagellate of microbacteria and Archea68. This family has a high diversity with multiple morphologies such as flagellated (Cercomonadidae, Glissomonadida) and spherical ameboid forms with filopodia (Vampyrellidae; Fig. S3), which could explain its high number of species found across all treatments69. Furthermore, lower temperatures for the duration of the experiment most likely aided their competitive ability. Sakharova and Korneva70 and Cruaud et al.71 found an increase of chrysophytes and Ciliphora during low temperatures in the Rybinsk Reservoir, Moscow and the Saint-Charles River, Canada suggesting these populations were active during low water temperatures. It could be hypothesised that the lower temperatures dampened the competitive ability of other algal groups and combined with nutrient limitation gave the SAR supergroup an advantage over the other phytoplankton groups resulting in their dominance in all treatments.
We observed a decline in both bacterial and eukaryotic species richness compared to the 2019 mesocosm experiment (according to the Shannon Diversity Index; Table S5). The DNA data (based on the PCoA plot; Fig. 5) indicated a significant difference between the bentophos and the other treatments, suggesting nutrient limitation specially of P is the main driver of biodiversity. Only the green algae concentrations showed a significant increase as the experiment progresses for all mesocosms regardless of treatment whilst cyanobacteria, diatom and cryptophytes concentrations either showed insignificant change or a decline in concentration over the four-week experiment (Fig. 3). Lürling et al.72 found that the mean growth rates of chlorophytes were significantly higher (0.62 day−1) than cyanobacteria (0.42 day−1) at 20 °C. This supports previous findings indicating that higher growth rates for chlorophytes under certain temperature regimes22. It could be suggested that low temperatures for the duration of this experiment dampened cyanobacterial growth giving chlorophytes a competitive advantage during this experiment. Moreover, other studies have found that chlorophytes dominate in hypertrophic lakes due to the fast growing chlorophytes having a competitive advantage over the slow growing cyanobacteria even when inorganic nutrient concentrations are low73. This could explain why the bentophos treatment saw a similar ending relative abundance (according to the metabarcoding data: Fig. 5) to the eutrophic treatment.
Conclusion
In summary, this experiment has shown that the use of the bentophos treatment causes significant differences in prokaryotic and eukaryotic communities resulting in a decline in cyanobacterial taxa and reduced biodiversity particularly within the eukaryotes. Unseasonably cold temperatures for the duration of the experiment most likely led to the disappearance of the inoculated strain of M. aeruginosa, so it was not possible to test how this adapted strain respond under nutrient limiting conditions. It would be interesting to test if the bentophos treatment still has an impact under enhanced temperature and if this causes a reduction in cyanobacteria. Undertaking mesocosms experiments under more favourable weather conditions may help determine if further adaption gives M. aeruginosa a competitive advantage under predicted future conditions. This study highlights how boosting the competitive advantage of other groups using bentophos and other P binding materials can be a potential mediation strategy against harmful cyanobacterial blooms.
Materials and methods
Biological material and growth conditions of the Microcystis strain (M11)
The M. aeruginosa strain (M11) used in this study was isolated from Lake Tașaul, Romania (44.35°N 28.61°E), a eutrophic freshwater lake, in the summer of 2018. The lake had an average water temperature of 22 °C. After collection, the samples were deposited as non-axenic cultures in the Collection of Cyanobacteria and Algae (AICB) at the Institute of Biological Research in Cluj-Napoca, Romania74. The phylogenetic identity of strain M11 was confirmed using the 16S rDNA gene amplified with specific primers75. The PCR fragments were sequenced by a third-party company (Macrogen Europe, Amsterdam, The Netherlands), who confirmed the strain was M. aeruginosa.
M11 was adapted for two years (from June 2018 to June 2020) in semi-batch conditions (corresponding to approx. 200 generations) in ultrapure water-based BG11 medium76 under controlled 16 h:8 h light:dark conditions provided by white LED lamps (25 μmol photon m−2 s−1). The strain was grown under two different temperature conditions, 22 °C (mean summer temperature) and 26 °C (the predicted temperature by 210077). Samples were grown in triplicates using 100 ml glass tubes. Each sample was bubbled daily with filtered atmospheric air (0.22 µm; Minisart, Sartorius, Göttingen, Germany) for 90 min (15 min aeration every 4 h). Frequent bubbling was needed to prevent cells gathering and causing self-shading.
Growth rate measurement and thermal reaction norms (TRNs)
During the two-year adaptation period, the growth rate (μ) of both ambient and heat-adapted M11 were measured at the beginning of the adaptation (June 2020, Month 0) as well as after 8, 16 and 24 months of cultivation at 22 °C and 26 °C. All cultures were tested in their original environment (22 °C) to compare whether long-term acclimatizationoccurred at 26 °C (as compared to 22 °C), to determine whether adapting M11 to 26 °C gave them a competitive advantage when grown under ambient temperature (22 °C) again. Therefore, the growth rate was only measured at 22 °C regardless of whether the strain was previously adapted at 22 °C or 26 °C. Before measuring all cultures were subsampled and diluted to the same starting density of OD600 = 0.1 (approximately 1.06 × 106 cell L−1). All samples were transferred to the control temperature (22 °C) for 14 days, with a growth rate measurement taken every second day (based on OD measurements) covering the exponential growth period. Growth rate measurements were stopped after 14 days because at this time most cultures had reached the stationary growth phase. The growth rate was calculated using the equation, μ = (ln Nd − ln N0)/d, where N0 and Nd represent culture density at the beginning and the end of each test, and d is the duration of the test in days. Number of divisions per day (Div.day−1) was calculated as follows: Div.day−1 = μ/ln2, while generation time (Gen.t) was calculated according to: Gen.t = 1/Div.day−1.
TRNs were completed to determine the optimal and lethal temperature for growth for both ambient and heat-adapted M11 to see if two years of adaptation to increased temperature resulted in a shift in its lethal temperature. Both heat-adapted M11 and ambient-adapted M11 were grown at a range of temperatures from 20 to 40 °C, with intervals of 2 °C. The growth rate was calculated using the same method as above.
Primer design and optimization PCR
The primers for qPCR were designed with FastPCR software78 using In silico Primer search option and targeted cyanobacterial sequences obtained from GenBank as the input. The specificity of the primers was checked by in silico PCR option from the same software and Primer-BLAST79. Primers for the reference rnpB gene was based on the bibliography search80,81. For each primer pair, the annealing temperature (Ta) was optimised through gradient PCR, in reaction mixtures (25 μl) containing: 0.4 μM of each primer, 12.5 μl of 2 × GoTaq® Green Master Mix (Promega), 1 μl cDNA and RNase/DNase-free water. The PCR profile was: 5 min at 95 °C followed by 35 cycles of 30 s at 95 °C, 30 s at 8 different temperatures near the Ta determined by the in silico PCR for each primer pair, 30 s at 72 °C and a final elongation step of 3 min at 72 °C. After optimisation, the size of the PCR products was checked by agarose gel electrophoresis performed by standard protocols, using 1 × TAE buffer and optimal Ta was determined for each primer pair (TableS6).
RNA extraction, cDNA synthesis and RT-qPCR
After each growth rate experiment (completed at Month 0, 8, 16 and 24) for the ambient and heat-adapted M11, the biomass was centrifuged (to remove the supernatant) and harvested for RNA and subsequent cDNA synthesis and RT-qPCR analysis to determine gene expression of seven thermal tolerance genes. For RNA extraction, the TRIzol® Reagent (Zymo Research) was used in combination with phenol–chloroform extraction. RNA was quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and samples were further treated with TURBO DNase treatment (Thermo Scientific) to remove any trace of genomic DNA contamination. For cDNA synthesis, 1 μg of total RNA was transcribed with the iScript™ Reverse Transcription Supermix (Bio-Rad Laboratories) in a final volume of 20 μl, following the manufacturer’s instructions.
The RT-qPCRs were carried out in triplicate on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using Eva Green detection chemistry. The reaction mixtures contained the following components: 5 μl 2 × Sso Fast EvaGreen SuperMix, 0.3 μM of the forward and reverse primers, 1 μl cDNA and RNase/DNase-free water to a final volume of 10 μl. The forward and reverse primers of the reference gene (rnpB) and target genes (cya1, pnp, pyrR, clpC1, clpC2 and sigF)25 were used to determine if these genes were expressed and if there was any difference between ambient and heat-adapted M11 (Table S6). The program consisted of an initial denaturation at 95 °C for 180 s, followed by 44 cycles of 15 s at 95 °C, an annealing step performed at 56 °C for 15 s and an elongation step at 72 °C for 30 s. For reaction specificity assessments, a post-PCR melting curve analysis was performed, in which the temperature ramped between 65 and 95 °C in 0.5 °C increments with subsequent plate readings. Amplification efficiencies of individual runs were checked with the LinRegPCR software. Outliers and samples diverging from the melting curve were omitted from further analysis. Fold change gene expression values of target and reference genes across different experimental conditions were calculated using the comparative Ct method (2−ΔΔCt)82.
Land-based mesocosm experiment
The impact of ambient and heat adapted M. aeruginosa strain, M11 on natural phytoplankton communities was tested at the Seeon Limnological Station (SLS) of the Ludwig-Maximilians-Universität (LMU) Munich, in Bavaria, Germany during a four-week mesocosm experiment (June-July 2020). Three 1000 L water tanks were used, in which 9 mesocosms were deployed consisting of translucent plastic (LDPE, Polyverpackung GmbH, Trappenkamp, Germany). The three different treatments used were oligotrophic lake water (Lake Klostersee, 48.08°N 11.96°E, TP < 10 mg/L), eutrophic lake water (Lake Bansee, 47.964°N 12.44°E, TP > 30 mg/L) and eutrophic lake water with the addition of bentophos (Dr. Nowak Institute, Ottersberg, Germany). Before the lake water was added to the tanks it was filtered (using a 250 µm gauze), to remove macro- and mesozooplankton. The temperature was not manipulated in any of the tanks (as the expected average July temperature was 23 °C), however a temperature measurement was collected every 15 min using sensors linked to a HOBO MX2202 temperature/light data logger. The three tanks were maintained in this way for 14 days, to allow the planktonic resident communities to acclimate to these conditions, prior to inoculation with M. aeruginosa and the addition of the bentophos to treatment 3. Then, the M. aeruginosa was inoculated into the lake communities, on June 26th 2020. The use of dialysis bags (10–20 k Dalton pore size; Nadir, Carl Roth, Karlsruhe, Germany), which are permeable for micro- and macro- nutrient, allowed three replicates of both ambient-adapted and heat-adapted M. aeruginosa for each treatment. Every enclosure was equipped with three replicate 1.4 L dialysis bags, each filled with water from their respective enclosures. Each dialysis bag was inoculated with strain M11 (initial concentration: 3 × 103 cells mL−1) for the controls, with both ambient and heat-adapted M11 being separately inoculated into all three treatments (identical to the methods used for the 2019 mesocosm experiment, as reported in Drugă et al.22. In total, the experiment consisted of 36 dialysis bags distributed into 9 enclosures, including the controls (dialysis bags with no added M. aeruginosa cells).
Chlorophyll-a concentration measurements
For the mesocosm experiment, chlorophyll a concentrations (µgL−1) were estimated in vivo using an AlgaeLabAnalyser device (bbe Moldaenke GmbH, Schwentinental, Germany) once a week, for a total of four weeks. The AlgaeLabAnalyser uses specific wavelengths to provide data on phytoplankton community composition based for four defined functional groups of total chlorophyll a concentrations. Four different major algal groups were identified: chlorophytes, cryptophytes, cyanobacteria and a brown group, which consists mainly of genera that have additional pigments that absorb in the yellow/orange wavelength range (diatoms, cryophytes, dinoflagellates).
Nutrients
Nutrient concentrations of nitrite (NO2), silicate (SiO2), particulate phosphorus (PO4-P), nitrate (NO3−) and total phosphorus (TP) concentrations were collected at the end of the four-week experiment for all treatments.
DNA analysis
The phytoplankton community composition of each dialysis bag was analysed after the four week mesocosm experiment using DNA metabarcoding. Firstly, the water from each of the three replicates was combined (totalling approx. 3 l), and then centrifuged to remove the excess water. Total DNA was isolated using a E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA), following the manufacturer’s instructions. The DNA concentration and quality were assessed using a NanoDrop™ 2000 Spectrophotometer (Waltham, MA, USA). An aliquot of the small ribosomal DNA (rDNA) subunit (16S for prokaryotes and 18S for eukaryotes) was then amplified using PCR with primers covering a wide range of planktonic taxa83,84. These primers allow for the amplification of a 450 bp DNA fragment within the 16S gene in prokaryotes, and a 650 bp long fragment from the V4 and V5 regions in the 18S gene in eukaryotes (Table S6). DNA sequencing was completed at the LMU Munich. In summary, 5 μL from each PCR reaction were pooled into separate 16S and 18S pools, purified with 0.8 × SPRI beads according to a standard protocol and eluted in 200 μL 10 mM Tris pH7.5. Libraries were constructed using the NEBNext UltraTMII DNA library preparation kit, and sequencing was done with a HiSeq 1500 system (Illumina, San Diego, CA, USA).
Following DNA sequencing, base calling and run demultiplexing were completed using the BaseSpace service (Illumina, San Diego, CA, USA) meters. The pair-end reads were joined in QIIME, and the quality filtration, dereplication and singleton removal was performed using Usearch v8. Both de novo and reference chimera checking were performed in Usearch v8, and using the last version of the Greengenes database (‘13_8’) as a reference85. The taxonomy was assigned for the representative OTUs in QIIME using the SSU/LSU 132 SILVA database86. The taxonomy was added to the OTU table with the biom-format package, and the mitochondrial and plastid sequences were filtered out of the final OTU table. Rarefaction was performed on 32,584 sequences, followed by alpha- and beta-diversity estimation in QIIME87,88. Visualisation of beta-diversity was performed using CANOCO 5 and R (Version 4.1.2).
Statistics (Data Analysis)
One-way repeated measures ANOVAs (on chlorophyll a concentrations) were completed for both ambient- and heat- adapted M11 together and for each version separately using the factor treatment. Prior to analysis the normality, the identification of outliers and assumption of sphericity were checked using a Shapiro Wilk’s test, “identifiy_outliers” function and Mauchly’s test of sphericity respectively. For the repeated measures ANOVAs, the control treatments were excluded as they contained no additional M. aeruginosa (and would distort the correlations). Post-hoc analysis included mean separation tests for the multiple comparisons (using Tukey-adjusted comparison) and least square means for the main effects. The analysis was completed using R (version 4.1.2).
Principal coordinate analysis (PCoA) was completed using the Weighted Unifrac of Bray–Curtis distances to determine the differences between treatments. This analysis was completed using the calibrate package in R (version 4.1.2). Principal component analysis (PCA) was conducted on the 16S and 18S metabarcoding data to summarise any groupings between the ambient and heat- adapted M11 and the treatments in an attempt to explain which species could describe these differences. It was completed at phylum and order level for both 16S and 18S genes using Canoco 5. Multiple alpha diversity indexes (Table S5) were used to estimate the diversity of the communities at the end of the experiment.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Material).
References
Dudgeon, D. et al. Freshwater biodiversity: Importance, threats, status and conservation challenges. Biol. Rev. Camb. Philos. Soc. 81, 163–182 (2006).
Grzybowski, M. & Glińska-Lewczuk, K. Principal threats to the conservation of freshwater habitats in the continental biogeographical region of Central Europe. Biodivers. Conserv. 28, 4065–4097 (2019).
Søndergaard, M. & Jeppesen, E. Anthropogenic impacts on lake and stream ecosystems, and approaches to restoration. J. Appl. Ecol. 44, 1089–1094 (2007).
Paerl, H. W., Fulton, R. S., Moisander, P. H. & Dyble, J. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. Sci. World J. 1, 76–113 (2001).
Krztoń, W., Kosiba, J., Pociecha, A. & Wilk-Woźniak, E. The effect of cyanobacterial blooms on bio- and functional diversity of zooplankton communities. Biodivers. Conserv. 28, 1815–1835 (2019).
Adrian, R. et al. Lakes as sentinels of climate change. Limnol. Oceanogr. 54, 2283–2297 (2009).
Dokulil, M. T. et al. Increasing maximum lake surface temperature under climate change. Clim. Change 165, 1–17 (2021).
Yan, X. et al. Climate warming and cyanobacteria blooms: Looks at their relationships from a new perspective. Water Res. 125, 449–457 (2017).
Paerl, H. W., Hall, N. S. & Calandrino, E. S. Controlling harmful cyanobacterial blooms in a world experiencing anthropogenic and climatic-induced change. Sci. Total Environ. 409, 1739–1745 (2011).
Anderson, D., Glibert, P. & Burkholder, J. Harmful algal blooms and eutrophication: Nutrient sources, compositions, and consequences. Estuaries 25, 704–726 (2002).
Li, D. et al. Factors associated with blooms of cyanobacteria in a large shallow lake, China. Environ. Sci. Eur. https://doi.org/10.1186/s12302-018-0152-2 (2018).
Rigosi, A., Carey, C. C., Ibelings, B. W. & Brookes, J. D. The interaction between climate warming and eutrophication to promote cyanobacteria is dependent on trophic state and varies among taxa. Limnol. Ocean. 59, 99–144 (2014).
Paerl, H. W. & Huisman, J. Blooms like it hot. Science 320, 57–58 (2008).
Paerl, H. W. Nuisance phytoplankton blooms in coastal, estuarine, and inland waters 1. Limnol. Oceanogr. 33, 823–843 (1988).
Schindler, D. W. et al. Eutrophication of lakes cannot be controlled by reducing nitrogen input: Results of a 37-year whole-ecosystem experiment. Proc. Natl. Acad. Sci. 105, 11254–11258 (2008).
Förster, W. et al. Phosphorous supply to a eutrophic artificial lake: Sedimentary versus groundwater sources. Water 13, 1–20 (2021).
Lang, P. et al. Phytoplankton community responses in a shallow lake following lanthanum-bentonite application. Water Res. 97, 55–68 (2016).
Lürling, M. & van Oosterhout, F. Case study on the efficacy of a lanthanum-enriched clay (Phoslock®) in controlling eutrophication in Lake Het Groene Eiland (The Netherlands). Hydrobiologia 710, 253–263 (2013).
Bishop, W. M. & Richardson, R. J. Influence of Phoslock® on legacy phosphorus, nutrient ratios, and algal assemblage composition in hypereutrophic water resources. Environ. Sci. Pollut. Res. 25, 4544–4557 (2018).
Drugă, B. et al. The impact of cation concentration on Microcystis (cyanobacteria) scum formation. Sci. Rep. 9, 1–11 (2019).
Stockenreiter, M., Isanta Navarro, J., Buchberger, F. & Stibor, H. Community shifts from eukaryote to cyanobacteria dominated phytoplankton: The role of mixing depth and light quality. Freshw. Biol. 66, 2145–2157 (2021).
Drugă, B. et al. term acclimation might enhance the growth and competitive ability of Microcystis aeruginosa in warm environments. Freshw. Biol. https://doi.org/10.1111/fwb.13865 (2022).
Fordham, D. A. Mesocosms reveal ecological surprises from climate change. PLOS Biol. 13, 1–7 (2015).
Reinl, K. L. et al. Cyanobacterial blooms in oligotrophic lakes: Shifting the high-nutrient paradigm. Freshw. Biol. 66, 1846–1859 (2021).
Tillich, U. M., Wolter, N., Franke, P., Dühring, U. & Frohme, M. Screening and genetic characterization of thermo-tolerant Synechocystis sp. PCC6803 strains created by adaptive evolution. BMC Biotechnol. 14, 1–15 (2014).
Burki, F., Roger, A. J., Brown, M. W. & Simpson, A. G. B. The new tree of eukaryotes. Trends Ecol. Evol. 35, 43–55 (2020).
LaPanse, A. J., Krishnan, A. & Posewitz, M. C. Adaptive Laboratory Evolution for algal strain improvement: Methodologies and applications. Algal Res. 53, 102122 (2021).
Deeg, C. M. et al. Chromulinavorax destructans, a pathogen of microzooplankton that provides a window into the enigmatic candidate phylum Dependentiae. PLOS Pathog. 15, e1007801 (2019).
Glöckner, F. O. et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. U. S. A. 100, 8298–8303 (2003).
Sowell, S. M. et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3, 93–105 (2009).
Chiriac, M.-C. et al. Ecogenomics sheds light on diverse lifestyle strategies in freshwater CPR. Microbiome https://doi.org/10.21203/rs.3.rs-776685/v2 (2022).
Von Der Heyden, S., Chao, E. E. & Cavalier-Smith, T. Genetic diversity of goniomonads: An ancient divergence between marine and freshwater species. Eur. J. Phycol. 39, 343–350 (2004).
Kim, B. R., Nakano, S. I., Kim, B. H. & Han, M. S. Grazing and growth of the heterotrophic flagellate Diphylleia rotans on the cyanobacterium Microcystis aeruginosa. Aquat. Microb. Ecol. 45, 163–170 (2006).
Varol, M., Bekleyen, A., Şen, B. & Gökot, B. First record of the order Choanoflagellida in Turkey. Turkish J. Fish. Aquat. Sci. 11, 1–2 (2011).
Cabrerizo, M. J. et al. Warming and CO2 effects under oligotrophication on temperate phytoplankton communities. Water Res. https://doi.org/10.1016/j.watres.2020.115579 (2020).
Maberly, S. C., Pitt, J.-A., Davies, P. S. & Carvalho, L. Nitrogen and phosphorus limitation and the management of small productive lakes. Inl. Waters 10, 159–172 (2020).
Li, J., Sellner, K., Place, A., Cornwell, J. & Gao, Y. Mitigation of cyanohabs using phoslock® to reduce water column phosphorus and nutrient release from sediment. Int. J. Environ. Res. Public Health https://doi.org/10.3390/ijerph182413360 (2021).
Nwosu, E. C. et al. Species-level spatio-temporal dynamics of cyanobacteria in a hard-water temperate lake in the southern Baltics. Front. Microbiol. 12, 1–17 (2021).
Vörös, L., Callieri, C., V-Balogh, K. & Bertoni, R. Freshwater picocyanobacteria along a trophic gradient and light quality range. Hydrobiologia 369–370, 117–125 (1998).
Camacho, A. On the occurrence and ecological features of deep chlorophyll maxima (DCM) in Spanish stratified lakes. Limnetica 25, 453–478 (2006).
Cabello-Yeves, P. J. et al. Novel synechococcus genomes reconstructed from freshwater reservoirs. Front. Microbiol. 8, 1151 (2017).
Prihantini, N. B., Addana, F., Sjamsuridzal, W. & Yokota, A. The effect of temperature on the growth of genus Synechococcus isolated from four Indonesian hot springs and Agathis small lake of Universitas Indonesia. AIP Conf. Proc. 1729 (2016).
Callieri, C. Synechococcus plasticity under environmental changes. FEMS Microbiol. Lett. 364, 1–8 (2017).
Acinas, S. G., Haverkamp, T. H. A., Huisman, J. & Stal, L. J. Phenotypic and genetic diversification of Pseudanabaena spp. (cyanobacteria). ISME J. 3, 31–46 (2009).
Kehoe, D. M. & Gutu, A. Responding to color: The regulation of complementary chromatic adaptation. Annu. Rev. Plant Biol. 57, 127–150 (2006).
Bell, T. & Kalff, J. The contribution of picophytoplankton in marine and freshwater systems of different trophic status and depth. Limnol. Oceanogr. 46, 1243–1248 (2001).
Jezberová, J. & Komárková, J. Morphological transformation in a freshwater Cyanobium sp. induced by grazers. Environ. Microbiol. 9, 1858–1862 (2007).
Chu, Z., Jin, X., Iwami, N. & Inamori, Y. The effect of temperature on growth characteristics and competitions of Microcystis aeruginosa and Oscillatoria mougeotii in a shallow, eutrophic lake simulator system. In Eutrophication of Shallow Lakes with Special Reference to Lake Taihu, China (eds Qin, B. et al.) 217–223 (Springer, 2007).
Ma, J. et al. The persistence of cyanobacterial (Microcystis spp,) blooms throughout winter in Lake Taihu, China. Limnol. Oceanogr. 61, 711–722 (2016).
Davis, T. W., Berry, D. L., Boyer, G. L. & Gobler, C. J. The effects of temperature and nutrients on the growth and dynamics of toxic and non-toxic strains of Microcystis during cyanobacteria blooms. Harmful Algae 8, 715–725 (2009).
Jankowiak, J., Hattenrath-Lehmann, T., Kramer, B. J., Ladds, M. & Gobler, C. J. Deciphering the effects of nitrogen, phosphorus, and temperature on cyanobacterial bloom intensification, diversity, and toxicity in western Lake Erie. Limnol. Oceanogr. 64, 1347–1370 (2019).
Martin, R. M. et al. Episodic decrease in temperature increases mcy gene transcription and cellular microcystin in continuous cultures of Microcystis aeruginosa PCC 7806. Front. Microbiol. 11, 3081 (2020).
You, J., Mallery, K., Hong, J. & Hondzo, M. Temperature effects on growth and buoyancy of Microcystis aeruginosa. J. Plankton Res. 40, 16–28 (2018).
Aguilar, P., Acosta, E., Dorador, C. & Sommaruga, R. Large differences in bacterial community composition among three nearby extreme waterbodies of the high Andean plateau. Front. Microbiol. 7, 1–8 (2016).
Echeverría-Vega, A. et al. Watershed-induced limnological and microbial status in two oligotrophic andean lakes exposed to the same climatic scenario. Front. Microbiol. 9, 1–17 (2018).
Schmidt, M. L., White, J. D. & Denef, V. J. Phylogenetic conservation of freshwater lake habitat preference varies between abundant bacterioplankton phyla. Environ. Microbiol. 18, 1212–1226 (2016).
Kaboré, O. D., Godreuil, S. & Drancourt, M. Planctomycetes as host-associated bacteria: A perspective that holds promise for their future isolations, by mimicking their native environmental niches in clinical microbiology laboratories. Front. Cell Infect. Microbiol. 10, 1–19 (2020).
Song, H., Li, Z., Du, B., Wang, G. & Ding, Y. Bacterial communities in sediments of the shallow Lake Dongping in China. J. Appl. Microbiol. 112, 79–89 (2012).
Sutcliffe, I. C. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470 (2010).
Zhang, W. et al. Phenotype changes of cyanobacterial and microbial distribution characteristics of surface sediments in different periods of cyanobacterial blooms in Taihu Lake. Aquat. Ecol. 54, 591–607 (2020).
Waidner, L. A. & Kirchman, D. L. Diversity and distribution of ecotypes of the aerobic anoxygenic phototrophy gene pufM in the Delaware estuary. Appl. Environ. Microbiol. 74, 4012–4021 (2008).
Sisson, C., Gulla-Devaney, B., Katz, L. A. & Grattepanche, J. D. Seed bank and seasonal patterns of the eukaryotic SAR (Stramenopila, Alveolata and Rhizaria) clade in a New England vernal pool. J. Plankton Res. 40, 376–390 (2018).
Moser, M. & Weisse, T. The outcome of competition between the two chrysomonads Ochromonas sp. and Poterioochromonas malhamensis depends on pH. Eur. J. Protistol. 47, 79–85 (2011).
Pröschold, T. et al. An integrative approach sheds new light onto the systematics and ecology of the widespread ciliate genus Coleps (Ciliophora, Prostomatea). Sci. Rep. 11, 1–19 (2021).
Jones, H. A classification of mixotrophic protists based on their behaviour. Freshw. Biol. 37, 35–43 (1997).
Fischer, R., Giebel, H. A. & Ptacnik, R. Identity of the limiting nutrient (N vs. P) affects the competitive success of mixotrophs. Mar. Ecol. Prog. Ser. 563, 51–63 (2017).
Gillette, J. P., Stewart, D. J., Teece, M. A. & Schulz, K. L. Abundance of mixoplanktonic algae in relation to prey, light and nutrient limitation in a dystrophic lake: A mesocosm study. Mar. Freshw. Res. 72, 1760–1772 (2021).
Harder, C. B. et al. Local diversity of heathland Cercozoa explored by in-depth sequencing. ISME J. 10, 2488–2497 (2016).
Ortiz-Álvarez, R., Triadó-Margarit, X., Camarero, L., Casamayor, E. O. & Catalan, J. High planktonic diversity in mountain lakes contains similar contributions of autotrophic, heterotrophic and parasitic eukaryotic life forms. Sci. Rep. 8, 1–12 (2018).
Sakharova, E. G. & Korneva, L. G. Phytoplankton in the Littoral and Pelagial zones of the Rybinsk reservoir in years with different temperature and water-level regimes. Inl. Water Biol. 11, 6–12 (2018).
Cruaud, P. et al. Annual Protist community dynamics in a freshwater ecosystem undergoing contrasted climatic conditions: The saint-Charles River (Canada). Front. Microbiol. https://doi.org/10.3389/fmicb.2019.02359 (2019).
Lürling, M., Eshetu, F., Faassen, E. J., Kosten, S. & Huszar, V. L. M. Comparison of cyanobacterial and green algal growth rates at different temperatures. Freshw. Biol. 58, 552–559 (2013).
Jensen, J. P., Jeppesen, E., Olrik, K. & Kristensen, P. Impact of nutrients and physical factors on the shift from cyanobacterial to chlorophyte dominance in shallow Danish lakes. Can. J. Fish. Aquat. Sci. 51, 1692–1699 (1994).
Dragoș, N. An Introduction to the Algae and the Culture Collection of Algae at the Institute of Biological Research, Cluj-Napoca (Cluj University Press, 1997).
Frank, J. A. et al. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470 (2008).
Allen, M. M. & Stanier, R. Y. Growth and division of some unicellular blue-green algae. J. Gen. Microbiol. 51, 199–202 (1968).
IPCC. IPCC report Global warming of 1.5°C. Ipcc Sr15 2, 17–20 (2018).
Kalendar, R., Khassenov, B., Ramankulov, Y., Samuilova, O. & Ivanov, K. I. FastPCR: An in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109, 312–319 (2017).
Ye, J. et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13, 1–11 (2012).
Kimura, S. et al. Diurnal infection patterns and impact of Microcystis cyanophages in a Japanese pond. Appl. Environ. Microbiol. 78, 5805–5811 (2012).
Pinto, F., Pacheco, C. C., Ferreira, D., Moradas-Ferreira, P. & Tamagnini, P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE 7, e34983 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408 (2001).
Hadziavdic, K. et al. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 9, e87624 (2014).
Herlemann, D. P. R. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579 (2011).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Lozupone, C. & Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Acknowledgements
This work was supported by Norway Grants Call 2019 (RO-NO 2019) –Collaborative Research Projects, no. 28/2020, and Romanian Ministry of Research, Innovation and Digitization (PN-III-P2-2.1-PED-2021, contract 653/2022 and the core programme PN2019–2022 –BIODIVERS 3, grant 25N/2019 –BIOSERV). It was also partly funded by a Transnational Access granted to BD through AQUACOSM EU Horizon 2020 INFRAIA-project No 731065 (CyanoWarm), by AQUACOSM-Plus (Horizon 2020-EU. 1.4.1.2, Grant agreement ID: 871081). Dr Andreas Brachman (from the Biocenter of the LMU Munich) is gratefully acknowledged for the technical assistance and the help concerning DNA sequencing.
Author information
Authors and Affiliations
Contributions
B.D. designed the experimental setup. E.S., B.D., and A.H performed the long-term adaptation experiment. E.S., M.S. and B.D. ran the mesocosm tests and chlorophyll analyses. M.N. measured the gene expression and thermal reaction norms, while C.L.B. and C.C analyzed the DNA data and performed the statistics. C.L.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Briddon, C.L., Szekeres, E., Hegedüs, A. et al. The combined impact of low temperatures and shifting phosphorus availability on the competitive ability of cyanobacteria. Sci Rep 12, 16409 (2022). https://doi.org/10.1038/s41598-022-20580-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20580-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.