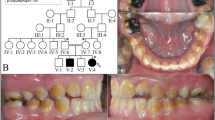
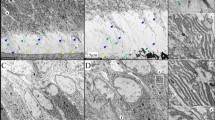
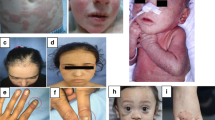
Abstract
Ameloblasts are specialized epithelial cells in the jaw that have an indispensable role in tooth enamel formation—amelogenesis1. Amelogenesis depends on multiple ameloblast-derived proteins that function as a scaffold for hydroxyapatite crystals. The loss of function of ameloblast-derived proteins results in a group of rare congenital disorders called amelogenesis imperfecta2. Defects in enamel formation are also found in patients with autoimmune polyglandular syndrome type-1 (APS-1), caused by AIRE deficiency3,4, and in patients diagnosed with coeliac disease5,6,7. However, the underlying mechanisms remain unclear. Here we show that the vast majority of patients with APS-1 and coeliac disease develop autoantibodies (mostly of the IgA isotype) against ameloblast-specific proteins, the expression of which is induced by AIRE in the thymus. This in turn results in a breakdown of central tolerance, and subsequent generation of corresponding autoantibodies that interfere with enamel formation. However, in coeliac disease, the generation of such autoantibodies seems to be driven by a breakdown of peripheral tolerance to intestinal antigens that are also expressed in enamel tissue. Both conditions are examples of a previously unidentified type of IgA-dependent autoimmune disorder that we collectively name autoimmune amelogenesis imperfecta.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The bulk RNA-seq dataset of mTECs isolated from NOD Aire+/+ and NOD Aire−/− mice is publicly available at the NCBI Gene Expression Omnibus (GEO) under accession number GSE224247. The remaining data are available within the Article and its Supplementary Information. Source data are provided with this paper.
References
Pham, C.-D. et al. Endocytosis and enamel formation. Front. Physiol. 8, 529 (2017).
Smith, C. E. L. et al. Amelogenesis imperfecta; genes, proteins, and pathways. Front. Physiol. 8, 435 (2017).
Pavlič, A. & Waltimo-Sirén, J. Clinical and microstructural aberrations of enamel of deciduous and permanent teeth in patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Arch. Oral Biol. 54, 658–665 (2009).
Bruserud, Ø. et al. A longitudinal follow-up of autoimmune polyendocrine syndrome type 1. J. Clin. Endocrinol. Metab. 101, 2975–2983 (2016).
Wierink, C. D., Van Diermen, D. E., Aartman, I. H. A. & Heymans, H. S. A. Dental enamel defects in children with coeliac disease. Int. J. Paediatr. Dent. 17, 163–168 (2007).
Bucci, P. et al. Oral aphthous ulcers and dental enamel defects in children with coeliac disease. Acta Paediatr. 95, 203–207 (2006).
Nieri, M., Tofani, E., Defraia, E., Giuntini, V. & Franchi, L. Enamel defects and aphthous stomatitis in celiac and healthy subjects: systematic review and meta-analysis of controlled studies. J. Dent. 65, 1–10 (2017).
Aschenbrenner, K. et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8, 351–358 (2007).
Wyss, L. et al. Affinity for self antigen selects Treg cells with distinct functional properties. Nat. Immunol. 17, 1093–1101 (2016).
Abramson, J. & Husebye, E. S. Autoimmune regulator and self-tolerance—molecular and clinical aspects. Immunol. Rev. 271, 127–140 (2016).
Peterson, P. & Peltonen, L. Autoimmune polyendocrinopathy syndrome type 1 (APS1) and AIRE gene: new views on molecular basis of autoimmunity. J. Autoimmun. 25, 49–55 (2005).
Pekka, A., Myllärniemi, S., Sipilä, I. & Perheentupa, J. Clinical variation of autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) in a series of 68 patients. N. Engl. J. Med. 322, 1829–1836 (1990).
Nishikawa, Y. et al. Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J. Exp. Med. 207, 963–971 (2010).
Vazquez, S. E. et al. Identification of novel, clinically correlated autoantigens in the monogenic autoimmune syndrome APS1 by proteome-wide PhIP-seq. eLife 9, e55053 (2020).
Catassi, C., Gatti, S. & Fasano, A. The new epidemiology of celiac disease. J. Pediatr. Gastroenterol. Nutr. 59, S7–S9 (2014).
Verbeek, W. H. M. et al. The spectrum of celiac disease: epidemiology, clinical aspects and treatment. Nat. Rev. Gastroenterol. Hepatol. 7, 204–213 (2010).
Dieterich, W. et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3, 797–801 (1997).
Sollid, L. M. Coeliac disease: dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2, 647–655 (2002).
Stenman, S. M. et al. Secretion of celiac disease autoantibodies after in vitro gliadin challenge is dependent on small-bowel mucosal transglutaminase 2-specific IgA deposits. BMC Immunol. 9, 6 (2008).
Jericho, H. & Guandalini, S. Extra-intestinal manifestation of celiac disease in children. Nutrients 10, 755 (2018).
Pastore, L. et al. Oral manifestations of celiac disease. J. Clin. Gastroenterol. PAP, 224–232 (2008).
Aine, L. Permanent tooth dental enamel defects leading to the diagnosis of coeliac disease. Br. Dent. J. 177, 253–254 (1994).
Petronijevic, S., Stig, S., Gao, J. & Halstensen, T. S. Amelogenin specific IgA and IgG in children with untreated coeliac disease. Eur. J. Oral Sci. 124, 526–533 (2016).
Mariani, P. et al. Coeliac disease, enamel defects and HLA typing. Acta Paediatr. 83, 1272–1275 (1994).
Pemberton, T. J. et al. Identification of novel genes expressed during mouse tooth development by microarray gene expression analysis. Dev. Dyn. 236, 2245–2257 (2007).
Eckstein, M. et al. Store-operated Ca2+ entry controls ameloblast cell function and enamel development. JCI Insight 2, e91166 (2017).
Duverger, O. et al. Hair keratin mutations in tooth enamel increase dental decay risk. J. Clin. Invest. 124, 5219–5224 (2014).
Chiba, Y. et al. G protein-coupled receptor Gpr115 (Adgrf4) is required for enamel mineralization mediated by ameloblasts. J. Biol. Chem. 295, 15328–15341 (2020).
Sharir, A. et al. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat. Cell Biol. 21, 1102–1112 (2019).
Sansom, S. N. et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 24, 1918–1931 (2014).
Bornstein, C. et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626 (2018).
Hu, J. C.-C. & Yamakoshi, Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit. Rev. Oral Biol. Med. 14, 387–398 (2003).
Nelson, S. J. Dental Anatomy, Physiology and Occlusion (Elsevier, 2015).
Perniola, R., Tamborrino, G., Marsigliante, S. & de Rinaldis, C. Assessment of enamel hypoplasia in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). J. Oral Pathol. Med. 27, 278–282 (1998).
Calamari, Z. T., Hu, J. K. H. & Klein, O. D. Tissue mechanical forces and evolutionary developmental changes act through space and time to shape tooth morphology and function. BioEssays 40, e1800140 (2018).
Katchburian, E., Katchburiant, A. V. & Pearse, A. G. E. Histochemistry of lysosomal enzymes in developing teeth of albino rats. J. Anat. 101, 783–792 (1967).
Baratella, L., Arana-Chavez, V. E. & Katchburian, E. Macrophages and apoptosis in the stellate reticulum of the rat enamel organ. J. Anat. 197, 303–306 (2000).
Nishikawa, S. & Sasaki, F. Phagocytotic processing of apoptotic bodies of transitional ameloblasts by MHC Class II-expressing macrophages in rat incisor. J. Histochem. Cytochem. 44, 1459–1467 (1996).
Tsuruga, E., Sakakura, Y., Yajima, T. & Shide, N. Appearance and distribution of dendritic cells and macrophages in dental pulp during early postnatal morphogenesis of mouse mandibular first molars. Histochem. Cell Biol. 112, 193–204 (1999).
Nishikawa, S. & Sasaki, F. Internalization of amelogenin by dendritic cells of the papillary layer during transition and early maturation stages. Histochem. Cell Biol. 112, 301–305 (1999).
Jessen, H. & Moe, H. The fine structure of macrophages in the enamel organ, with special reference to the microtubular system. Z. Zellforsch. 126, 466–182 (1972).
Bossù, M., Bartoli, A., Orsini, G., Luppino, E. & Polimeni, A. Enamel hypoplasia in coeliac children: a potential clinical marker of early diagnosis. Eur. J. Paediatr. Dent. 8, 31–37 (2007).
Petronijevic, S., Stig, S. & Halstensen, T. S. Epitope mapping of anti‐amelogenin IgA in coeliac disease. Eur. J. Oral Sci. 128, 27–36 (2020).
Sóñora, C. et al. Enamel organ proteins as targets for antibodies in celiac disease: implications for oral health. Eur. J. Oral Sci. 124, 11–16 (2016).
Paolella, G., Sposito, S., Romanelli, A. M. & Caputo, I. Type 2 transglutaminase in coeliac disease: a key player in pathogenesis, diagnosis and therapy. Int. J. Mol. Sci. 23, 7513 (2022).
Herrera, M. G. & Dodero, V. I. Gliadin proteolytical resistant peptides: the interplay between structure and self-assembly in gluten-related disorders. Biophys. Rev. 13, 1147–1154 (2021).
Kristjánsson, G., Venge, P. & Hällgren, R. Mucosal reactivity to cow’s milk protein in coeliac disease. Clin. Exp. Immunol. 147, 449–455 (2007).
Capone, K., Sansotta, N., Vohra, P., Jericho, H. & Guandalini, S. Milk protein-induced villous atrophy and elevated serologies in four children with celiac disease on a gluten-free diet. Ann. Pediatr. 3, 1028 (2020).
Coucke, F. Food intolerance in patients with manifest autoimmunity. Observational study. Autoimmun. Rev. 17, 1078–1080 (2018).
Zone, J. J., Egan, C. A., Taylor, T. B. & Meyer, L. J. IgA autoimmune disorders: development of a passive transfer mouse model. J. Invest. Dermatol. Symp. Proc. 9, 47–51 (2004).
Sinnberg, T. et al. Pulmonary surfactant proteins are inhibited by immunoglobulin A autoantibodies in severe COVID-19. Am. J. Respir. Crit. Care Med. 207, 38–49 (2023).
Mylliirniemi, S. & Perheentupa, J. Oral findings in the autoimmune polyendocrinopathy-candidosis syndrome (APECS) and other forms of hypoparathyroidism. Oral Surg. Oral Med. Oral Pathol. 45, 721–729 (1978).
Rashid, M. & Zarkadas, M. Oral manifestations of celiac disease: a clinical guide for dentists. J. Can. Dent. Assoc. 77, b39 (2011).
Gibbons, D. L. & Spencer, J. Mouse and human intestinal immunity: same ballpark, different players; different rules, same score. Mucosal Immunol. 4, 148–157 (2011).
Monteiro, R. C. & Van De Winkel, J. G. J. IgA Fc receptors. Annu. Rev. Immunol. 21, 177–204 (2003).
Laible, G., Smolenski, G., Wheeler, T. & Brophy, B. Increased gene dosage for β- and κ-casein in transgenic cattle improves milk composition through complex effects. Sci. Rep. 6, 37607 (2016).
Bijl, E., van Valenberg, H. J. F., Huppertz, T. & van Hooijdonk, A. C. M. Protein, casein, and micellar salts in milk: current content and historical perspectives. J. Dairy Sci. 96, 5455–5464 (2013).
Wedholm, A., Larsen, L. B., Lindmark-Månsson, H., Karlsson, A. H. & Andrén, A. Effect of protein composition on the cheese-making properties of milk from individual dairy cows. J. Dairy Sci. 89, 3296–3305 (2006).
Smith, C. E. L. et al. Phenotype and variant spectrum in the LAMB3 form of amelogenesis imperfecta. J. Dent. Res. 98, 698–704 (2019).
Kim, J.-W. et al. A novel de novo mutation in LAMB3 causes localized hypoplastic enamel in the molar region. Eur. J. Oral Sci. 124, 403–405 (2016).
Poulter, J. A. et al. Whole-exome sequencing, without prior linkage, identifies a mutation in LAMB3 as a cause of dominant hypoplastic amelogenesis imperfecta. Eur. J. Hum. Genet. 22, 132–135 (2013).
Caccamo, D. et al. Expression pattern of transglutaminases in the early differentiation stage of erupting rat incisor. Amino Acids 36, 49–56 (2009).
Beth, S. A. et al. Generation R birth cohort study shows that specific enamel defects were not associated with elevated serum transglutaminase type 2 antibodies. Acta Paediatr. 105, e485–e491 (2016).
McCarra, C., Olegário, I. C., O’Connell, A. C. & Leith, R. Prevalence of hypomineralised second primary molars (HSPM): a systematic review and meta-analysis. Int. J. Paediatr. Dent. 32, 367–382 (2022).
Kohen, R. et al. UTAP: user-friendly transcriptome analysis pipeline. BMC Bioinform. 20, 154 (2019).
Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Xu, H. et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157 (2015).
Concordet, J. P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
De Laurenzi, V. & Melino, G. Gene disruption of tissue transglutaminase. Mol. Cell. Biol. 21, 148–155 (2001).
Jiang, W., Anderson, M. S., Bronson, R., Mathis, D. & Benoist, C. Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 202, 805–815 (2005).
Ossart, J. et al. Breakdown of immune tolerance in AIRE-deficient rats induces a severe autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy-like autoimmune disease. J. Immunol. 201, 874–887 (2018).
Vera Alvarez, R., Pongor, L. S., Mariño-Ramírez, L. & Landsman, D. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 35, 1960–1962 (2019).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Khan, F., He, M. & Taussig, M. J. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on Ni-nitrilotriacetic acid surfaces. Anal. Chem. 78, 3072–3079 (2006).
Wald, T. et al. Intrinsically disordered enamel matrix protein ameloblastin forms ribbon-like supramolecular structures via an N-terminal segment encoded by exon 5. J. Biol. Chem. 288, 22333–22345 (2013).
Simmer, J. P. et al. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif. Tissue Int. 54, 312–319 (1994).
Wilkinson, D. G. & Nieto, M. A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225, 361–373 (1993).
Goldfarb, Y. et al. Mechanistic dissection of dominant AIRE mutations in mouse models reveals AIRE autoregulation. J. Exp. Med. 218, e20201076 (2021).
Acknowledgements
Research in the Abramson laboratory is supported by the European Research Council (ERC-2016-CoG-724821), the Bill and Marika Glied and Family Fund, the Israel Science Foundation (1819/21), the Binational Science Foundation (BSF) and the Joseph and Sarah Bollag Fund. J.A. is an incumbent of the Eugene and Marcia Applebaum Professorial Chair and of the IOCB fellowship for sabbatical visit program (RVO 61388963). Y. Gruper is supported by Ariane de Rothschild PhD fellowship. E.S.H. was supported by The Weston Visiting Professorship program at The Weizmann Institute of Science. Additional sources of support were provided by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR) projects LM2023053 (to R.O.), LM2018126, LM2023036 and Upgrade of the Czech Centre for Phenogenomics CZ.02.1.01/0.0/0.0/16_013/0001789 and CZ.02.1.01/0.0/0.0/18_046/0015861 by MEYS and ESIF (to R. Sedlacek), Czech Science Foundation, GACR 19-19025Y (to J.P.); Czech Academy of Science, CAS RVO 6837805 0(toR.S.); and the KG Jebsen Center for Autoimmune Disorders and The Research Council of Norway and the Western Norway Health Authorities and the University of Bergen (to A.S.B.W. and E.S.H.); and the National Research, Development and Innovation Office, NKFI, Hungary (K138162) (to Z.S.). We thank the staff at the BIOCEV Imaging Methods Core Facility (supported by MEYS CR LM2018129 and ERDF CZ.02.1.01/0.0/0.0/18_046/0016045), the Center of Molecular Structure CMS-Biocev (supported by MEYS CR LM2018127) and S. Kozubová for technical help, and P. B. Juliusson at the University of Bergen for providing healthy control sera from children.
Author information
Authors and Affiliations
Contributions
J.A., E.S.H. and A.S.B.W. conceived and coordinated the study. Unless stated otherwise, J.A., E.S.H. and Y. Gruper planned and designed the experiments and analyses in this study. Unless specifically stated, experiments and analyses were performed by Y. Gruper. A.S.B.W. performed part of the ELISA experiments with APS-1 sera. L.G. generated the Csn3−/− mice and performed part of the qPCR, ELISA and IF experiments, and analysed together with Y. Gruper the μCT data. F.S. performed μCT and SEM analyses for NOD Aire−/− and WT mice. Y.H. cloned mouse-ameloblast-specific genes and performed some initial western blotting and bioinformatic analyses. Y. Goldfarb generated RNA-seq datasets for mTECs isolated from NOD mice. G.A.-N. performed Csn3 RNA-ISH expression analysis and contributed to immunohistochemistry and IF data. A.O., T.W. and R.O. designed and constructed expression vectors and produced and purified recombinant proteins. J.D., N.K., T. Gome, E.F. and S.B.-D. contributed to data analyses. O.B.-N., A.B., B.L., T. Givony, R.K. and C.S. assisted in some experiments. B.M. produced recombinant enamelin. E.S.H., E.M.O. and M.C.M. examined patients with APS-1. M.B. and C.G. provided sera from Aire−/− rats. K.E.A.L., R. Shamir, D.S.S., N.T. and E.D. examined patients with coeliac disease and provided the patients’ sera. J.P. isolated mouse ameloblasts and supervised μCT, immunohistochemistry, ISH and SEM analyses and ENAM production. R. Sedlacek contributed to μCT, ISH and SEM experiments and subsequent data interpretation. S.F. and T.P. performed the histology staining for TGM2. C.H. and I.L. collected data for TGM2 μCT analysis and performed SEM analysis of TGM2 enamel. Z.S. coordinated the TGM2-related studies and analysed and interpreted the data. J.A. supervised the study. J.A. and Y. Gruper wrote the manuscript and A.S.B.W., E.S.H., R.O., J.P., C.G., Y. Goldfarb, J.D., Y.H., Z.S., R. Shamir and K.E.A.L. critically revised it.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Detlef Schuppan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 AIRE induces ameloblast genes in mTECs.
a, Illustration of the main amelogenesis developmental stages. The inductive (pre-secretory) stage (1) gives rise to functional secretory ameloblasts (2), which secrete various enamel matrix proteins, thereby providing a scaffold for hydroxyapatite crystal formation. Next, in the maturation stage (3), the secreted enamel matrix proteins undergo proteolytic degradation by enamel matrix specific enzymes. This consequently gives rise to the hardest mineralized tissue in the body, the enamel. Created with BioRender.com; b, Illustration of two alternative mechanisms responsible for enamel dysplasia in AIRE-deficient individuals. Created with BioRender.com; c,d, Reanalysis of published microarray data sets25 showing differential gene expression of mouse developing teeth relative to other mouse tissues. Differential gene expression was compared to gene’s mean expression (c), or to the corresponding p-value of genes (d), (Fc>2.0, Log2ME > 4, p-value < 0.05). Amelogenesis imperfecta linked genes (red), tooth-specific genes (blue) and Aire (black). e, Reanalysis of published scRNA-seq dataset29 showing expression of the indicated genes in dental epithelium; high expression (green), no expression (black). f, Heatmap showing normalized expression values (as log2) of amelogenesis imperfecta linked genes and Aire in tooth germ and ameloblast (determined by RNA-seq analyses26,27,28). g, Relative gene expression of Aire and Amelx in ameloblast cells (at E15.5, 5 days, 3 weeks and 16 weeks) and in mTEChi (at 4 weeks) as assayed by qPCR. h,i, Representative immunofluorescence microscopy images of NOD WT (h) or NOD Aire−/− (i) jaw and thymus sections stained with AIRE-specific antibodies (green), amelogenin-specific antibodies (red) and DAPI (blue). Scale bar 100 µm. j, Bulk RNA-seq results of normalized expression of amelogenesis imperfecta linked genes (red), tooth specific genes (blue) and AIRE-depended positive control gene (black) in NOD WT (n = 3) or NOD Aire−/− (n = 3) mTECs. Raw P values were adjusted for multiple testing, using the procedure of Benjamini and Hochberg65. k,l, Volcano plots of bulk RNA-seq of ameloblast-specific genes (blue), amelogenesis imperfecta linked genes (red) or AIRE-depended positive control gene (black) in mTEChi cells isolated from either NOD (k) or B630 (l) Aire−/− versus WT mice. In bold are AIRE-dependent genes depicted in mice of both backgrounds. n refers to the number of biologically independent mice per genotype or group. Data are representative of at least two independent experiments. Data shown as mean ± s.e.m. Data were analysed using one-way ANOVA following Tukey’s multiple comparisons test (g).
Extended Data Fig. 2 AIRE-deficient mice develop autoantibodies against a battery of ameloblast-specific antigens.
a-e, UMAP of published31 scRNA-seq of thymic epithelial cells (TEC; EpCAM+CD45–) from B6 WT mice. Each dot represents a single cell; a, UMAP highlighting the presence of five major colour-coded TEC clusters. b, UMAP of Aire gene expression per TEC subset as assayed by scRNA-seq. c,d, UMAPs of ameloblast-specific and amelogenesis imperfecta related genes whose expression is primarily restricted (c) or not restricted (d) to the Aire+ TEC subset. e, UMAPs of ameloblast-specific and amelogenesis imperfecta related genes with undetectable expression levels within the TEC compartment as assayed by scRNA-seq. f, Representative immunofluorescence microscopy images of Rag1−/− mouse jaw sections stained with sera (green) from NOD Aire−/− mice (upper panel) or WT mice (lower panel), amelogenin-specific antibodies (red) and DAPI (blue). Scale bar 100 µm. g, Slot-blot analysis of primary ameloblast tissue lysate probed with commercial antibodies to the indicated ameloblast-specific proteins. h, Slot-blot analysis measuring the reactivity of sera from NOD Aire−/− (n ≥ 9) or WT mice (n ≥ 5) to the indicated ameloblast-specific recombinant proteins. Upper panel shows mouse IgG/M sera reactivity against ameloblast-specific protein. Lower panel shows the same membrane blotted with commercial antibodies specific to the corresponding recombinant protein, or ponceau staining. Each lane represents an individual mouse. i, ELISA analyses measuring the reactivity of IgG/M specific to the indicated ameloblast-specific proteins in sera from NOD Aire−/− (n = 14, n = 12 for ODAM and FAM20A) or NOD WT mice (n = 14, n = 13 for FAM20A, n = 9 for ODAM). j, ELISA analyses measuring the reactivity of IgA specific to the indicated ameloblast-specific proteins in sera from NOD Aire−/− (n = 9) or NOD WT mice (n = 6). k, ELISA analyses showing distribution of IgG/M sera reactivity of female vs male in NOD Aire−/− (n = 14; M = 7, F = 7) or WT mice (n = 14; M = 7, F = 7). n refers to the number of biologically independent mice per genotype or group. Data are representative of at least three independent experiments. Data shown as mean ± s.e.m. Data were analysed using two-sided Fisher exact test (i,j) and two-way ANOVA following Bonferroni post-tests (k).
Extended Data Fig. 3 AIRE-deficient rats and patients with APS-1 develop autoantibodies against a battery of ameloblast-specific antigens.
a, Slot-blot analyses measuring the reactivity of sera from Aire−/− (n ≥ 8) or WT rats (n ≥ 5) to the indicated ameloblast-specific recombinant proteins. Upper panel shows rat IgG sera reactivity against ameloblast-specific protein. Lower panel shows the same membrane blotted with commercial antibodies specific to the corresponding recombinant protein, or ponceau staining. Each lane represents an individual rat. b,c, ELISA analyses measuring the reactivity of IgG (b) or IgA (c) specific to the indicated ameloblast-specific recombinant proteins in sera from Aire−/− (n ≥ 15) or WT (n ≥ 5) rats. d, Representative immunofluorescence microscopy images of Rag1−/− mouse jaw sections stained with sera (green) from paediatric patients with APS-1 (upper panel) or healthy control (lower panel), with LAMB3-specific antibodies (red), and DAPI (blue). Scale bar 50 µm. e–g, ELISA analyses measuring the reactivity of IgA/IgG1 specific to the indicated ameloblast-specific proteins in sera from adult (e and g) or paediatric (f) patients with APS-1 (adults n = 22, paediatric n = 17) or healthy controls (adults n = 20, paediatric n = 18). n refers to the number of biologically independent rats/patients per genotype or group. Data are representative of at least three independent experiments. Data shown as mean ± s.e.m. Data were analysed using two-sided Fisher exact test (b,c, e–g).
Extended Data Fig. 4 Defective enamel formation presented in AIRE-deficient mice and linked to enamel-specific autoantibody levels in patients with APS-1.
a, ELISA analyses measuring the reactivity of IgG2, IgG3 and IgG4 antibodies in sera isolated from adult patients with APS-1 (n = 10) or healthy controls (n = 5) to the indicated ameloblast-specific recombinant proteins. b, ELISA analyses showing distribution of IgA/IgG1 sera reactivity of adult females versus males in patients with APS-1 (Females n = 11; Males n = 12, n = 11 for ENAM). c, Violin plots showing enamel defects score distribution of each tooth type (Maxillary teeth) of 4–29-year-old patients with APS-1 (n = 14). Red line represents median value. d, Representation of enamel calcification duration (in years) for each tooth type. e, Correlation between the duration of enamel calcification in years (Molars=3 y, Incisors=4.7 y, Canines=6.5 y) versus the mean enamel defects score for each tooth type (Molars=1.25, Incisors=1.8, Canines=2.43) (in Maxillary teeth) of paediatric patients with APS-1 (n = 14). f, Comparison of gross teeth appearance in 14–24-week-old NOD Aire−/− and WT mice. Representative images of ~45 mice teeth examined. g, Heatmap showing distribution of enamel area (mm2) within the left and right (L&R) incisors of the individual NOD Aire−/− (n = 12) and WT (n = 11) mice. The length of the individual incisors was normalized, and the specific relative position on the tooth displayed in percentiles (y axis). The heatmap range was defined individually per tooth to present the largest (red), median (white) and smallest (blue) enamel area per tooth. h, Presentation of enamel mineralization (a.u.) along the relative (%) incisor location (from lateral to apical part) in representative NOD Aire−/− (n = 12) and WT (n = 11) mice. Vertical dotted lines indicate a region corresponding to the apical part (that is the 75th−95th percentile of the incisor’s length). n refers to the number of biologically independent mice/patients per genotype or group. Data are representative of at least two independent experiments. Data shown as mean ± s.e.m. Data were analysed using two-sided Fisher exact test (a), two-sided unpaired t-test (b), one-way ANOVA following Tukey’s multiple comparisons test (c) and two-sided Pearson R correlation test (e).
Extended Data Fig. 5 Defective enamel formation in AIRE-deficient mice is driven by autoantibodies.
a, Illustration of two alternative autoreactive mechanisms responsible for defective enamel matrix formation. The response mediated by self-reactive T-cells (left) would result in the destruction of the ameloblast layer (in an analogous manner to the autoimmune destruction of β-cells in type 1 diabetes or retinal degeneration in Aire−/− mice80), while the autoantibody-mediated response (right) would interfere with enamel matrix formation (e.g. through blocking functional epitopes on enamel matrix proteins), rather than cause destruction of ameloblasts. Created with BioRender.com. b, Representative H&E staining images of jaws isolated from NOD Aire−/− and WT mice showing enamel matrix and ameloblast cells in secretory stage at a proximal and distal orientation. Red arrows indicate immune cell infiltrates. Left and middle images scale bar 50 µm, right images scale bar 20 µm. c, Representative immunohistochemical analysis images of CD8+ T-cells, F4.80+ macrophages, CD11c+ Dendritic cells and CD20+ B-cells within the ameloblast layer, in teeth sections from NOD Aire−/− and Aire+/− mice. Scale bar 50 µm. d, Representative immunofluorescence microscopy image of CX3CR1-GFP mouse jaw section stained with amelogenin-specific antibodies (red) and DAPI (blue). Scale bar 20 µm. e, Representative immunofluorescence microscopy image of NOD Aire−/− mouse jaw section stained with MHC-II (RT1B)-specific antibodies (green), amelogenin-specific antibodies (red) and DAPI (blue). Scale bar 100 µm. f, Representative immunofluorescence microscopy images of CX3CR1-GFP mouse jaw section stained with amelogenin-specific antibodies (red), MHC-II (I-A/I-E)-specific antibodies (purple) and DAPI (blue). Scale bar 20 µm. g, Illustration of the serum transfer experimental design shown in h–k. Created with BioRender.com. h, Percentage of mean variability in enamel mineralization at the indicated apical part between left and right incisors of WT mice transferred with sera isolated either from aged NOD Aire−/− (red) (n = 6) or WT (blue) (n = 5) mice in two independent experimental cohorts. i, Presentation of enamel mineralization (a.u.) along the relative (%) incisor location (from lateral to apical part) in WT mice transferred with sera isolated either from aged NOD Aire−/− (n = 6) (red and orange) or WT (n = 5) (dark and light blue) mice. Vertical dotted lines indicate a region corresponding to the apical part (that is the 75th−95th percentile of the incisor’s length). Red arrows highlight areas with high variability between left and right teeth. Presentation of two independent transfer experiments. j,k, Representative SEM images showing ultrastructure of enamel matrix of teeth isolated from WT mice that were injected with sera isolated from NOD WT (j) or NOD Aire−/− (k) mice, as illustrated in g. Scale bar 20 µm. l, Illustration of the proposed model for ameloblast-specific autoantibody production in patients with APS-1: as a result of AIRE deficiency and break of central tolerance, autoreactive T-cells escape from the thymus to the periphery (1). The cells circulate the body and enter lymph nodes (2). In parallel, during the formation of the enamel matrix, antigen presenting cells (APC) sample enamel antigens (3) and migrate to the closest draining lymph node (4), where they present the enamel self-antigens to autoreactive CD4+ T-cells that later activate autoreactive B-cells to produce antibodies against enamel self-antigens (5). The plasma cells and/or autoantibodies are then transferred via the bloodstream to the dental area, where the enamel matrix is developing, bind the enamel matrix proteins and thereby interfere with their capacity to aid in hydroxyapatite mineralization and deposition and enamel formation (6). Illustration was created with BioRender.com. n refers to the number of biologically independent mice per genotype or group. Data are representative of at least two independent experiments. Data shown as mean ± s.e.m. Data were analysed using two-sided Mann Whitney test (h).
Extended Data Fig. 6 Patients with coeliac disease develop IgA autoantibodies to ameloblast-specific proteins and intestinal antigens.
a, ELISA analyses measuring the reactivity of IgA specific to the indicated enamel proteins in sera from paediatric patients with coeliac disease (n = 24, n = 23 for AMBN) or control individuals without coeliac disease (n = 21). b, ELISA analyses measuring the reactivity of IgA/IgG1 specific to the indicated enamel proteins in sera from adult patients with coeliac disease (n = 48, n = 39 for AMBN and AMTN) or healthy controls (n = 14, n = 48 for FAM20A, n = 6 for AMBN and AMTN). c, Correlation between IgA reactivity to TGM2 vs IgA reactivity to the indicated enamel proteins in paediatric patients with coeliac disease (n = 24) (black dots) and control individuals without coeliac disease (n = 21) (blue dots). d, Representative immunofluorescence microscopy images of Rag1−/− mouse jaw sections stained with sera (green) from patients with coeliac disease (upper panel) or healthy control (lower panel), amelogenin-specific antibodies (red) and DAPI (blue). Scale bar 50 µm. e, ELISA analysis measuring the titre levels of TGM2-specific-sera (IgA) isolated from paediatric patients with coeliac disease (n = 24) or control individuals without coeliac disease (n = 21). f, Representative immunofluorescence microscopy images of Rag1−/− mouse small intestine sections stained with sera (green) from patients with coeliac disease (upper panel) or healthy control (lower panel), TGM2-specific antibodies (red) and DAPI (blue). Scale bar 50 µm. g, Diaminobenzidine (DAB) staining with anti-TGM2 specific antibody showing localization of TGM2 in the developing tooth (IEE=inner enamel epithelium; PreSecA= presecretory ameloblast; SecA=secretory ameloblast) of WT animal. Scale bar 50 µm (upper panel), 25 µM (middle panel) and 20 µM (lower panel). h, Representative immunofluorescence microscopy images of Rag1−/− mouse jaw sections stained with sera (green) from patients with coeliac disease (upper panel) or healthy control (lower panel), TGM2-specific antibodies (red) and DAPI (blue). Scale bar 50 µm. i, Presentation of enamel area (mm2; upper panel) or mineralization (a.u.; lower panel) along the relative (%) incisor location (from lateral to apical part) in representative Tgm2−/− (n = 4) (orange) and WT (n = 4) (blue) mice. Vertical dotted lines indicate a region corresponding to the apical part (that is, the 75th−95th percentile of the incisor’s length). j, Relative expression of LAMB3 gene in different human tissues, based on scRNA-seq datasets publicly available at the human protein atlas database. k, Representative immunofluorescence microscopy images of Rag1−/− mouse small intestine sections stained with sera (green) isolated from patients with coeliac disease celiac (upper panel) or healthy control (lower panel), laminin beta subunit 3-specific antibodies (red), TGM2-specific antibodies (yellow) and DAPI (blue). Scale bar 50 µm. n refers to the number of biologically independent mice/patients per genotype or group. Data are representative of at least two independent experiments. Data shown as mean ± s.e.m. Data were analysed using two-sided Fisher exact test (a, b and e), two-sided Pearson R correlation test (c).
Extended Data Fig. 7 κ-casein is a dental antigen.
a, Heatmap showing normalized expression values (as log2) of Csn3 in tooth germ and ameloblast (determined by RNA-seq analysis26,27,28) in comparison to known amelogenesis imperfecta linked genes, other casein genes and Aire. b, Relative gene expression of indicated ameloblast specific genes and Csn3 in neonatal jaw, salivary gland and liver as assayed by qPCR (n = 6 biologically independent animals). c, RNA-ISH analysis of Csn3 mRNA expression and localization in a mouse jaw. Longitudinal sections of mouse incisors where different stages of ameloblast differentiation are visible. Right panel staining with sense probe as a negative control. Left panel, staining with Csn3 probe. Black arrow indicates the position of the ameloblasts layer. Red arrow indicates the position of the odontoblasts layer. Scale bar 200 µm. d, Illustration of the Csn3 locus and the CRISPR design used for deleting Csn3 exon 1. DNA represented as a black line, with exon location indicated as grey frames. The location of the guide RNAs is indicated in pink frames. Primers for DNA sequencing are shown with arrows representing the amplification product of each reaction. Red filled area represent sequenced DNA. Illustration Created with SnapGene software 6.1 (GSL Biotech). Bottom, alignment of Csn3+/+ and Csn3−/− mice DNA sequence at the Csn3 gene locus. Exons are marked in yellow frames. e, Illustration of the PCR genotyping strategy of Csn3−/− mice. Illustration was created with BioRender.com; on the right is PCR analysis of Csn3+/+ (WT), Csn3−/+ (het) and Csn3−/− (KO) mice. Primer A, hybridizing upstream to exon 1 of Csn3, in combination with primer B results in amplification of a 276-bp fragment indicative of the WT Csn3 locus. Combination of primer A with primer C yields a 433-bp fragment specific for the knockout Csn3 locus, or 2436-bp fragment that cannot be seen in the gel and is specific to the WT Csn3 locus. f, Relative gene expression of Csn3 gene in neonatal jaw of Csn3−/− and WT mice as assayed by qPCR. Data are representative of at least two independent experiments. Data shown as mean ± s.e.m. Data were analysed using one-way ANOVA following Tukey’s multiple comparisons test for each gene separately (b), and two-sided unpaired t-test (f).
Extended Data Fig. 8 Defective enamel formation in CSN3-deficient mice and autoantibodies to κ-casein in patients with coeliac disease.
a,b, Presentation of enamel area (mm2) (a) or mineralization (a.u.) (b) along the relative (%) incisor location (from lateral to apical part) in representative Csn3−/− (n = 7) (orange) and WT (n = 7) (blue) mice. Vertical dotted lines indicate a region corresponding to the the apical part (that is, the 75th−95th percentile of the incisor’s length). c,d, Percentage of mean variability in enamel area (mm2) (c) or mineralization (a.u.) (d) at the indicated apical part between left and right incisors of Csn3−/− (red) (n = 7) or WT (blue) (n = 7) mice. e, Masks of sections from molar teeth roots (M1, M2 and M3) of WT (upper panel) or Csn3−/− (lower panel) mice. Root M2 marked by yellow frame. Structure abnormalities marked by red arrows. f, Representative images of µCT analysis on molars of Csn3−/− or WT mice, assessing dentine root thickness (upper panel) and pulp root thickness (lower panel) in pseudo colours. Images are of sagittal sections. h, ELISA analyses measuring the reactivity of IgA (g) or IgG1 (h) specific to bovine CSN3 in sera from paediatric patients with coeliac disease (n = 24) or control individuals without coeliac disease (n = 21). i, Correlation between reactivity of IgA vs IgG1 antibodies specific to bovine CSN3 in paediatric patients with coeliac disease (n = 24), as measured by ELISA. j, ELISA analysis measuring the reactivity of IgG1 specific to bovine CSN3 in sera isolated from adult patients with coeliac disease (n = 48) or healthy controls (n = 14). k, Correlation between reactivity of IgA vs IgG1 antibodies specific to bovine CSN3 in adult patients with coeliac disease (n = 48), as measured by ELISA. l, Amino-acid alignment of bovine and human CSN3 proteins. Domain structure of CSN3 protein after cleavage: Signal peptide (blue), para-kappa-casein (orange) and macro-peptide (green). Highly conserved epitopes between the bovine and human homologues are marked in red frames. m,n, ELISA analyses measuring the reactivity of IgA (m) or IgG1 (n) specific to human CSN3 in sera from paediatric patients with coeliac disease (n = 24) or control individuals without coeliac disease (n = 21). o, ELISA analysis measuring the reactivity of IgG1 specific to human CSN3 in sera from adult patients with coeliac disease (n = 48) or healthy controls (n = 14). p, Correlation between reactivity of IgA antibodies specific to bovine versus human CSN3 in paediatric patients with coeliac disease (n = 24), as measured by ELISA. q, ELISA analysis measuring the reactivity of IgA sera obtained from adult patients with coeliac disease (n = 48) or healthy controls (n = 14) towards a library of 25 amino-acid long peptides derived from human CSN3. r, Heatmap showing positive (red) or negative (blue) serum reactivity to the indicated proteins in paediatric patients with coeliac disease (n = 24) and control individuals without coeliac disease (n = 21), based on the ELISA results in Fig. 3a and Extended Data Fig. 6a. Each row represents a patient. s,t, ELISA analyses measuring the reactivity of IgA specific to human (s) or bovine (t) CSN3 in sera isolated from paediatric patients with APS-1 (n = 17) or healthy controls (n = 18). u, Normalized expression of Csn3 gene in WT or AIRE-deficient mTECs isolated from NOD mice. Raw P values were adjusted for multiple testing, using the procedure of Benjamini and Hochberg65. n refers to the number of biologically independent mice/patients per genotype or group. Data are representative of at least two independent experiments. Data shown as mean ± s.e.m. Data were analysed using two-sided Mann Whitney test (c and d), two-sided Fisher exact test (g, h, j, m, n, o, s and t), two-sided Pearson R correlation test (i, k and p), and two-way ANOVA followed by Bonferroni multiple comparisons test (q).
Extended Data Fig. 9 Model for ameloblast-specific autoantibody production in patients with coeliac disease.
Illustration of the proposed model for ameloblast-specific autoantibody production in patients with coeliac disease. The inflamed environment of the small intestine in patients with coeliac disease disrupts intestinal barrier capacity and increases intestinal permeability (i.e., leaky gut). This results in generation of antibodies to various food antigens with high immunogenicity (e.g., gliadins or caseins). Reactivity to food antigens is often accompanied by epitope and antigen spreading and subsequent production of autoantibodies to various proteins expressed in the intestinal microenvironment (e.g., TGM2 and LAMB3) (1). The plasma cells and/or autoantibodies are then transferred via the bloodstream to the developing enamel matrix where they can bind to their targets expressed in the dental tissues and enamel matrix (2). The opsonized enamel autoantigens can then be internalized by dental macrophages (and possibly other resident phagocytes), which serve as local sentinels and bind these autoantibodies via their FCGR/FCAR receptors expressed on the surface of these cells (3). Such interactions facilitate the internalization of the antibody-self-antigen complexes by the phagocytes (4). The engulfed antigens will then be presented by the phagocytes on their MHCII molecules in the context of inflammation to self-reactive T cell escapees that are normally kept in check by peripheral mechanisms of self-tolerance. This further potentiates the formation of the adaptive immune response, including further antibody formation and epitope spreading to enamel-specific proteins, in a self-perpetuating mechanism. Illustration was created with BioRender.com.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Supplementary Tables 1–9.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gruper, Y., Wolff, A.S.B., Glanz, L. et al. Autoimmune amelogenesis imperfecta in patients with APS-1 and coeliac disease. Nature 624, 653–662 (2023). https://doi.org/10.1038/s41586-023-06776-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06776-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.