Plasma Membrane Protein Nce102 Modulates Morphology and Function of the Yeast Vacuole
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Plasmids
2.3. Preparation of Freeze Fracture Replicas and Electron Microscopy
2.4. Vacuole Staining
2.5. Confocal Microscopy
2.6. Vacuole Isolation
2.7. Western Blot Analysis
3. Results
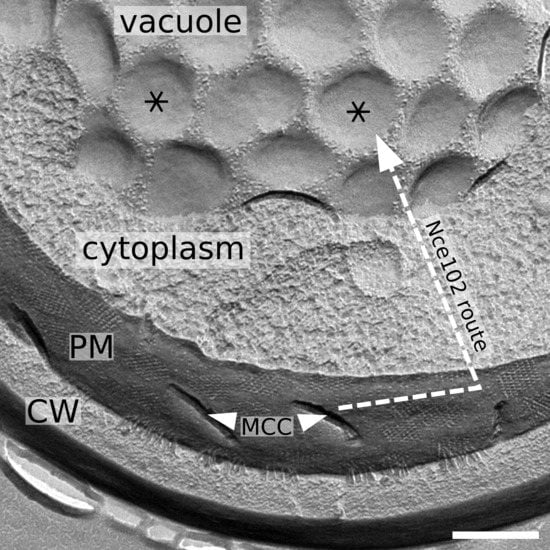
3.1. Nce102 Localizes to Ergosterol-Rich Microdomains of the Vacuolar Membrane
3.2. Vacuolar Fraction of Nce102 Originates in the Plasma Membrane
3.3. Nce102 Regulates Vacuolar Fusion and V-ATPase Stability
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grossmann, G.; Malinsky, J.; Stahlschmidt, W.; Loibl, M.; Weig-Meckl, I.; Frommer, W.B.; Opekarová, M.; Tanner, W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 2008, 183, 1075–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gournas, C.; Prévost, M.; Krammer, E.; André, B. Function and Regulation of Fungal Amino Acid Transporters: Insights from Predicted Structure. In Yeast Membrane Transport; Ramos, J., Sychrová, H., Kschischo, M., Eds.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Germany, 2016; Volume 892, pp. 69–106. ISBN 9783319253022. [Google Scholar]
- Busto, J.V.; Elting, A.; Haase, D.; Spira, F.; Kuhlman, J.; Schäfer-Herte, M.; Wedlich-Söldner, R. Lateral plasma membrane compartmentalization links protein function and turnover. EMBO J. 2018, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kubalová, D.; Káňovičová, P.; Veselá, P.; Awadová, T.; Džugasová, V.; Daum, G.; Malínský, J.; Balážová, M. The lipid droplet protein Pgc1 controls the subcellular distribution of phosphatidylglycerol. FEMS Yeast Res. 2019, 19, foz045. [Google Scholar] [CrossRef] [PubMed]
- Grousl, T.; Opekarová, M.; Stradalova, V.; Hasek, J.; Malinsky, J. Evolutionarily conserved 5′-3′ exoribonuclease Xrn1 accumulates at plasma membrane-associated eisosomes in post-diauxic yeast. PLoS ONE 2015, 10, e0122770. [Google Scholar] [CrossRef]
- Vaškovičová, K.; Awadová, T.; Veselá, P.; Balážová, M.; Opekarová, M.; Malinsky, J. mRNA decay is regulated via sequestration of the conserved 5′-3′ exoribonuclease Xrn1 at eisosome in yeast. Eur. J. Cell Biol. 2017, 96, 591–599. [Google Scholar] [CrossRef]
- Berchtold, D.; Piccolis, M.; Chiaruttini, N.; Riezman, I.; Riezman, H.; Roux, A.; Walther, T.C.; Loewith, R. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 2012, 14, 542–547. [Google Scholar] [CrossRef]
- Niles, B.J.; Mogri, H.; Hill, A.; Vlahakis, A.; Powers, T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. USA 2012, 109, 1536–1541. [Google Scholar] [CrossRef] [Green Version]
- Cleves, A.E.; Cooper, D.N.W.; Barondes, S.H.; Kelly, R.B. A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 1996, 133, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, F.; Moreira, K.; Aguilar, P.S.; Hubner, N.C.; Mann, M.; Walter, P.; Walther, T.C. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 2009, 185, 1227–1242. [Google Scholar] [CrossRef] [Green Version]
- García-Marqués, S.; Randez-Gil, F.; Dupont, S.; Garre, E.; Prieto, J.A. Sng1 associates with Nce102 to regulate the yeast Pkh–Ypk signalling module in response to sphingolipid status. BBA Mol. Cell Res. 2016, 1863, 1319–1333. [Google Scholar] [CrossRef]
- Malinsky, J.; Opekarová, M. New Insight Into the Roles of Membrane Microdomains in Physiological Activities of Fungal Cells; Academic Press: Cambridge, MA, USA, 2016; Volume 325, ISBN 9780128048061. [Google Scholar]
- Zahumensky, J.; Malinsky, J. Role of MCC/eisosome in fungal lipid homeostasis. Biomolecules 2019, 9, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loibl, M.; Grossmann, G.; Stradalova, V.; Klingl, A.; Rachel, R.; Tanner, W.; Malinsky, J.; Opekarová, M. C terminus of Nce102 determines the structure and function of microdomains in the Saccharomyces cerevisiae plasma membrane. Eukaryot. Cell 2010, 9, 1184–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabeche, R.; Baldissard, S.; Hammond, J.; Howard, L.; Moseley, J.B. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol. Biol. Cell 2011, 22, 4059–4067. [Google Scholar] [CrossRef] [PubMed]
- Athanasopoulos, A.; Gournas, C.; Amillis, S.; Sophianopoulou, V. Characterization of AnNce102 and its role in eisosome stability and sphingolipid biosynthesis. Sci. Rep. 2015, 5, 15200. [Google Scholar] [CrossRef]
- Stradalova, V.; Stahlschmidt, W.; Grossmann, G.; Blazikova, M.; Rachel, R.; Tanner, W.; Malinsky, J. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 2009, 122, 2887–2894. [Google Scholar] [CrossRef] [Green Version]
- Moeller, C.H.; Thomson, W.W. An ultrastructural study of the yeast tonoplast during the shift from exponential to stationary phase. J. Ultrastruct. Res. 1979, 68, 28–37. [Google Scholar] [CrossRef]
- Toulmay, A.; Prinz, W.A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 2013, 202, 35–44. [Google Scholar] [CrossRef]
- Malínská, K.; Malínský, J.; Opekarová, M.; Tanner, W. Visualization of Protein Compartmentation within the Plasma Membrane of Living Yeast Cells. Mol. Biol. Cell 2003, 14, 4427–4436. [Google Scholar] [CrossRef]
- Malinska, K.; Malinsky, J.; Opekarova, M.; Tanner, W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004, 117, 6031–6041. [Google Scholar] [CrossRef] [Green Version]
- Grossmann, G.; Opekarová, M.; Malinsky, J.; Weig-Meckl, I.; Tanner, W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 2007, 26, 1–8. [Google Scholar] [CrossRef]
- Rachel, R.; Wyschkony, I.; Riehl, S.; Huber, H. The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 2002, 1, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Wiederhold, E.; Gandhi, T.; Permentier, H.P.; Breitling, R.; Poolman, B.; Slotboom, D.J. The yeast vacuolar membrane proteome. Mol. Cell. Proteom. 2009, 8, 380–392. [Google Scholar] [CrossRef] [Green Version]
- Bevis, B.J.; Hammond, A.T.; Reinke, C.A.; Glick, B.S. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 2002, 4, 750–756. [Google Scholar] [CrossRef]
- Wang, C.W.; Miao, Y.H.; Chang, Y.S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 2014, 206, 357–366. [Google Scholar] [CrossRef] [Green Version]
- DeRisi, J.L.; Iyer, V.R.; Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997, 278, 680–686. [Google Scholar] [CrossRef] [Green Version]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Shapira, M.; Regev, A.; Pe’er, D.; Botstein, D.; Koller, D.; Friedman, N. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003, 34, 166–176. [Google Scholar] [CrossRef]
- Gournas, C.; Gkionis, S.; Carquin, M.; Twyffels, L.; Tyteca, D.; André, B. Conformation-dependent partitioning of yeast nutrient transporters into starvation-protective membrane domains -Supplement. Proc. Natl. Acad. Sci. USA 2018, 115, E3145–E3154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babst, M.; Wendland, B.; Estepa, E.J.; Emr, S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998, 17, 2982–2993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thayer, N.H.; Leverich, C.K.; Fitzgibbon, M.P.; Nelson, Z.W.; Henderson, K.A.; Gafken, P.R.; Hsu, J.J.; Gottschling, D.E. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc. Natl. Acad. Sci. USA 2014, 111, 14019–14026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.E.; Karpova, T.S.; Brugger, B.; Moschenross, D.M.; Wang, G.K.; Schneiter, R.; Wieland, F.T.; Cooper, J.A. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 2002, 22, 927–934. [Google Scholar] [CrossRef] [Green Version]
- Malinsky, J.; Opekarová, M.; Grossmann, G.; Tanner, W. Membrane Microdomains, Rafts, and Detergent-Resistant Membranes in Plants and Fungi. Annu. Rev. Plant Biol. 2013, 64, 501–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggi, M.; Kusmider, B.; Loewith, R. The flipside of the TOR coin—TORC2 and plasma membrane homeostasis at a glance. J. Cell Sci. 2020, 133, jcs242040. [Google Scholar] [CrossRef] [PubMed]
- Gournas, C.; Saliba, E.; Krammer, E.-M.; Barthelemy, C.; Prévost, M.; André, B. Transition of yeast Can1 transporter to the inward-facing state unveils an α-arrestin target sequence promoting its ubiquitylation and endocytosis. Mol. Biol. Cell 2017, 28, 2819–2832. [Google Scholar] [CrossRef]
- Bianchi, F.; Syga, L.; Moiset, G.; Spakman, D.; Schavemaker, P.E.; Punter, C.M.; Seinen, A.-B.B.; Van Oijen, A.M.; Robinson, A.; Poolman, B.; et al. Steric exclusion and protein conformation determine the localization of plasma membrane transporters. Nat. Commun. 2018, 9, 501. [Google Scholar] [CrossRef]
- Zhang, L.B.; Tang, L.; Guan, Y.; Feng, M.G. Subcellular localization of Sur7 and its pleiotropic effect on cell wall integrity, multiple stress responses, and virulence of Beauveria bassiana. Appl. Microbiol. Biotechnol. 2020, 104, 6669–6678. [Google Scholar] [CrossRef]
- Dawaliby, R.; Mayer, A. Microautophagy of the Nucleus Coincides with a Vacuolar Diffusion Barrier at Nuclear–Vacuolar Junctions. Mol. Biol. Cell 2010, 21, 4173–4183. [Google Scholar] [CrossRef] [Green Version]
- Levine, T.P.; Munro, S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 2001, 12, 1633–1644. [Google Scholar] [CrossRef] [Green Version]
- Murley, A.; Sarsam, R.D.; Toulmay, A.; Yamada, J.; Prinz, W.A.; Nunnari, J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 2015, 209, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manik, M.K.; Yang, H.; Tong, J.; Im, Y.J. Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction. Structure 2017, 25, 617–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasanthakumar, T.; Bueler, S.A.; Wu, D.; Beilsten-Edmands, V.; Robinson, C.V.; Rubinstein, J.L. Structural comparison of the vacuolar and Golgi V-ATPases from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2019, 116, 7272–7277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brett, C.L.; Kallay, L.; Hua, Z.; Green, R.; Chyou, A.; Zhang, Y.; Graham, T.R.; Donowitz, M.; Rao, R. Genome-Wide Analysis Reveals the Vacuolar pH-Stat of Saccharomyces cerevisiae. PLoS ONE 2011, 6, e17619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaigg, B.; Timischl, B.; Corbino, L.; Schneiter, R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J. Biol. Chem. 2005, 280, 22515–22522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, K.; Tanaka, T.; Furusawa, C.; Nagahisa, K.; Hirasawa, T.; Shimizu, H. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 32–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, J.L.; Srinivasan, B.; Dickson, R.C.; Lester, R.L. Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J. Bacteriol. 1992, 174, 7180–7184. [Google Scholar] [CrossRef] [Green Version]
- Malinsky, J.; Tanner, W.; Opekarova, M. Transmembrane voltage: Potential to induce lateral microdomains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 806–811. [Google Scholar] [CrossRef]
- Michaillat, L.; Mayer, A. Identification of Genes Affecting Vacuole Membrane Fragmentation in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e54160. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Pulido, L.; Martín-Belmonte, F.; Valencia, A.; Alonso, M.A. MARVEL: A conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002, 27, 599–601. [Google Scholar] [CrossRef]
- Te Welscher, Y.M.; Jones, L.; Van Leeuwen, M.R.; Dijksterhuis, J.; De Kruijff, B.; Eitzen, G.; Breukink, E. Natamycin inhibits vacuole fusion at the priming phase via a specific interaction with ergosterol. Antimicrob. Agents Chemother. 2010, 54, 2618–2625. [Google Scholar] [CrossRef] [Green Version]
- Audhya, A.; Loewith, R.; Parsons, A.B.; Gao, L.; Tabuchi, M.; Zhou, H.; Boone, C.; Hall, M.N.; Emr, S.D. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004, 23, 3747–3757. [Google Scholar] [CrossRef] [Green Version]
- Appadurai, D.; Gay, L.; Moharir, A.; Lang, M.J.; Duncan, M.C.; Schmidt, O.; Teis, D.; Vu, T.N.; Silva, M.; Jorgensen, E.M.; et al. Plasma membrane tension regulates eisosome structure and function. Mol. Biol. Cell 2020, 31, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Tafur, L.; Kefauver, J.; Loewith, R. Structural Insights into TOR Signaling. Genes (Basel) 2020, 11, 885. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, E. Rag GTPase in amino acid signaling. Amino Acids 2016, 48, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Zanolari, B.; Friant, S.; Funato, K.; Suetterlin, C.; Stevenson, B.J.; Riezman, H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2824–2833. [Google Scholar] [CrossRef]
- Friant, S. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 2000, 19, 2834–2844. [Google Scholar] [CrossRef] [Green Version]
- Lester, R.L.; Wells, G.B.; Oxford, G.; Dickson, R.C. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J. Biol. Chem. 1993, 268, 845–856. [Google Scholar]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaskovicova, K.; Vesela, P.; Zahumensky, J.; Folkova, D.; Balazova, M.; Malinsky, J. Plasma Membrane Protein Nce102 Modulates Morphology and Function of the Yeast Vacuole. Biomolecules 2020, 10, 1476. https://doi.org/10.3390/biom10111476
Vaskovicova K, Vesela P, Zahumensky J, Folkova D, Balazova M, Malinsky J. Plasma Membrane Protein Nce102 Modulates Morphology and Function of the Yeast Vacuole. Biomolecules. 2020; 10(11):1476. https://doi.org/10.3390/biom10111476
Chicago/Turabian StyleVaskovicova, Katarina, Petra Vesela, Jakub Zahumensky, Dagmar Folkova, Maria Balazova, and Jan Malinsky. 2020. "Plasma Membrane Protein Nce102 Modulates Morphology and Function of the Yeast Vacuole" Biomolecules 10, no. 11: 1476. https://doi.org/10.3390/biom10111476