Acute Pneumonia Caused by Clinically Isolated Legionella pneumophila Sg 1, ST 62: Host Responses and Pathologies in Mice
Abstract
:1. Introduction
2. Material and Methods
2.1. Mice
2.2. Bacteria Identification and Cultivation
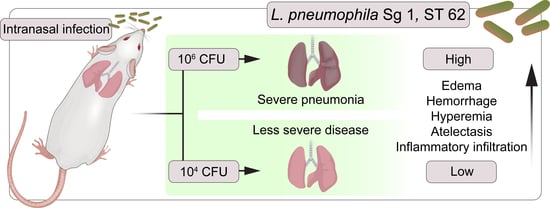
2.3. Mice Infection
2.4. Hematological Analysis
2.5. Histopathological Examination
2.6. Data Analysis
3. Results
3.1. Case Report and Isolate Characterization
3.2. Body Weight and Bacterial Burden
3.3. Hematological Analysis
3.4. Gross Pathology Appearance and Histopathological Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Watkins, K. Emerging Infectious Diseases: A Review. Curr. Emerg. Hosp. Med. Rep. 2018, 6, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Ellwanger, J.H.; Kaminski, V.d.L.; Chies, J.A.B. Emerging infectious disease prevention: Where should we invest our resources and efforts? J. Infect. Public Health 2019, 12, 313–316. [Google Scholar] [CrossRef] [PubMed]
- McClure, E.E.; Chávez, A.S.O.; Shaw, D.K.; Carlyon, J.A.; Ganta, R.R.; Noh, S.M.; Wood, D.O.; Bavoil, P.M.; Brayton, K.A.; Martinez, J.J.; et al. Engineering of obligate intracellular bacteria: Progress, challenges and paradigms. Nat. Rev. Microbiol. 2017, 15, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.F.; Hales, S.; White, P.S.; Baker, M.G. Review Global seroprevalence of legionellosis - a systematic review and meta-analysis. Sci. Rep. 2020, 10, 7337. [Google Scholar] [CrossRef]
- Chitasombat, M.N.; Ratchatanawin, N.; Visessiri, Y. Disseminated extrapulmonary Legionella pneumophila infection presenting with panniculitis: Sase report and literature review. BMC Infect. Dis. 2018, 18, 467. [Google Scholar] [CrossRef] [PubMed]
- Burke, P.T.; Shah, R.; Thabolingam, R.; Saba, S. Suspected Legionella-induced perimyocarditis in an adult in the absence of pneumonia: A rare clinical entity. Tex. Heart Inst. J. 2009, 36, 601–603, PMCID:PMC2801935. [Google Scholar]
- Bodur, H.; Savran, Y.; Koca, U.; Kilinç, O.; Albayrak, S.; Itil, O.; Akoglu, S. Legionella pneumonia with acute respiratory distress syndrome, myocarditis and septic shock successfully treated with Drotrecogin Alpha (activated). Eur. J. Anaesthesiol. 2006, 23, 808–810. [Google Scholar] [CrossRef]
- Sommer, J.B.; Erbguth, F.J.; Neundörfer, B. Acute disseminated encephalomyelitis following Legionella pneumophila infection. Eur. Neurol. 2000, 44, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, N.; Suzuki, H.; Tokuda, Y.; Takano, T. Severe Legionnaires’ disease with pneumonia and biopsy-confirmed myocarditis most likely caused by Legionella pneumophila serogroup 6. Intern. Med. 2012, 51, 3207–3212. [Google Scholar] [CrossRef] [Green Version]
- Dietert, K.; Gutbier, B.; Wienhold, S.M.; Reppe, K.; Jiang, X.; Yao, L.; Chaput, C.; Naujoks, J.; Brack, M.; Kupke, A.; et al. Spectrum of pathogen- and model-specific histopathologies in mouse models of acute pneumonia. PLoS ONE 2017, 12, e0188251. [Google Scholar] [CrossRef]
- Sabrià, M.; Campins, M. Legionnaires’ Disease. Am. J. Respir. Med. 2003, 2, 235–243. [Google Scholar] [CrossRef]
- Li, L.-H.; Zhang, L.; Wu, H.-Y.; Qu, P.-H.; Chen, J.-C.; Zhan, X.-Y.; Zhu, Q.-Y.; Chen, C.; Hu, C.-H. Legionella septentrionalis sp. nov., isolated from aquatic environments in the northern PR China. Int. J. Syst. Evol. Microbiol. 2021, 71, 1–7. [Google Scholar] [CrossRef]
- Yu, V.L.; Plouffe, J.F.; Pastoris, M.C.; Stout, J.E.; Schousboe, M.; Widmer, A.; Summersgill, J.; File, T.; Heath, C.M.; Paterson, D.L.; et al. Distribution of Legionella Species and Serogroups Isolated by Culture in Patients with Sporadic Community-Acquired Legionellosis: An International Collaborative Survey. J. Infect. Dis. 2002, 186, 127–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaber, L.; Amro, M.; Tair, H.A.; Bahader, S.A.; Alalam, H.; Butmeh, S.; Hilal, D.A.; Brettar, I.; Höfle, M.G.; Bitar, D.M. Comparison of in situ sequence type analysis of Legionella pneumophila in respiratory tract secretions and environmental samples of a hospital in East Jerusalem. Epidemiol. Infect. 2018, 146, 2116–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underwood, A.P.; Jones, G.; Mentasti, M.; Fry, N.K.; Harrison, T.G. Comparison of the Legionella pneumophilapopulation structure as determined by sequence-based typing and whole genome sequencing. BMC Microbiol. 2013, 13, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helbig, J.H.; Bernander, S.; Castellani Pastoris, M.; Etienne, J.; Gaia, V.; Lauwers, S.; Lindsay, D.; Lück, P.C.; Marques, T.; Mentula, S.; et al. Pan-European study on culture-proven Legionnaires’ disease: Distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, K.; Jacobs, E.; Röske, I.; Helbig, J.H. Legionella pneumophila carrying the virulence-associated lipopolysaccharide epitope possesses two functionally different LPS components. Microbiology (Read. Engl.) 2010, 156, 2953–2961. [Google Scholar] [CrossRef] [Green Version]
- Harrison, T.G.; Afshar, B.; Doshi, N.; Fry, N.K.; Lee, J.V. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000-2008). Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Kozak, N.A.; Benson, R.F.; Brown, E.; Alexander, N.T.; Taylor, T.H., Jr.; Shelton, B.G.; Fields, B.S. Distribution of lag-1 alleles and sequence-based types among Legionella pneumophila serogroup 1 clinical and environmental isolates in the United States. J. Clin. Microbiol. 2009, 47, 2525–2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, S.; Mentasti, M.; Tewolde, R.; Aslett, M.; Harris, S.R.; Afshar, B.; Underwood, A.; Fry, N.K.; Parkhill, J.; Harrison, T.G. Evaluation of an Optimal Epidemiological Typing Scheme for Legionella pneumophila with Whole-Genome Sequence Data Using Validation Guidelines. J. Clin. Microbiol. 2016, 54, 2135–2148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brieland, J.; Freeman, P.; Kunkel, R.; Chrisp, C.; Hurley, M.; Fantone, J.; Engleberg, C. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires’ disease. Am. J. Pathol. 1994, 145, 1537–1546, PMCID:PMC1887509. [Google Scholar] [PubMed]
- Brieland, J.; Remick, D.; Freeman, P.; Hurley, M.; Fantone, J.; Engleberg, C. In vivo regulation of replicative Legionella pneumophila lung infection by endogenous tumor necrosis factor alpha and nitric oxide. Infect. Imunn. 1995, 63, 3253–3258. [Google Scholar] [CrossRef] [Green Version]
- Brieland, J.K.; Loebenberg, D.; Menzel, F.; Hare, R.S. Efficacy of SCH27899 in an Animal Model of Legionnaires’; Disease Using Immunocompromised A/J Mice. Antimicrob. Agents Chemother. 2000, 44, 1333–1336. [Google Scholar] [CrossRef] [Green Version]
- Edelstein, P.H.; Calarco, K.; Yasui, V.K. Antimicrobial therapy of experimentally induced Legionnaires’ disease in guinea pigs. Am. Rev. Respir. Dis. 1984, 130, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, G.N.; Petty, N.K.; Mousnier, A.; Harding, C.R.; Vogrin, A.J.; Wee, B.; Fry, N.K.; Harrison, T.G.; Newton, H.J.; Thomson, N.R.; et al. Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J. Bacteriol. 2010, 192, 6001–6016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawn, T.R.; Berrington, W.R.; Smith, I.A.; Uematsu, S.; Akira, S.; Aderem, A.; Smith, K.D.; Skerrett, S.J. Altered inflammatory responses in TLR5-deficient mice infected with Legionella pneumophila. J. Immunol. 2007, 179, 6981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelstein, P.H.; Edelstein, M.A. In vitro activity of the ketolide HMR 3647 (RU 6647) for Legionella spp., its pharmacokinetics in guinea pigs, and use of the drug to treat guinea pigs with Legionella pneumophila pneumonia. Antimicrob. Agents. Chemother. 1999, 43, 90–95. [Google Scholar] [CrossRef] [Green Version]
- Davis, G.S.; Winn, W.C., Jr.; Gump, D.W.; Craighead, J.E.; Beaty, H.N. Legionnaires’ pneumonia after aerosol exposure in guinea pigs and rats. Am. Rev. Respir. Dis. 1982, 126, 1050–1057. [Google Scholar] [CrossRef]
- Helbig, J.H.; Kurtz, J.B.; Pastoris, M.C.; Pelaz, C.; Lück, P.C. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: Possibilities and limitations for division of the species into serogroups. J. Clin. Microbiol. 1997, 35, 2841–2845. [Google Scholar] [CrossRef] [Green Version]
- Qin, T.; Zhou, H.; Ren, H.; Guan, H.; Li, M.; Zhu, B.; Shao, Z. Distribution of sequence-based types of legionella pneumophila serogroup 1 strains isolated from cooling towers, hot springs, and potable water systems in China. Appl. Environ. Microbiol. 2014, 80, 2150–2157. [Google Scholar] [CrossRef] [Green Version]
- Fontana, S.; Scaturro, M.; Rota, M.C.; Caporali, M.G.; Ricci, M.L. Molecular typing of Legionella pneumophila serogroup 1 clinical strains isolated in Italy. Int. J. Med. Microbiol. 2014, 304, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Hori, J.I.; Zamboni, D.S. The mouse as a model for pulmonary legionella infection. Methods Mol. Biol. 2013, 954, 493–503. [Google Scholar] [CrossRef]
- Rayamajhi, M.; Delgado, C.; Condon, T.; Riches, D.; Lenz, L. Lung B cells promote early pathogen dissemination and hasten death from inhalation anthrax. Mucosal Immunol. 2012, 5, 444–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudel, S.; Baral, P.; Ghimire, L.; Bergeron, S.; Jin, L.; DeCorte, J.A.; Le, J.T.; Cai, S.; Jeyaseelan, S. CXCL1 regulates neutrophil homeostasis in pneumonia-derived sepsis caused by Streptococcus pneumoniae serotype 3. Blood 2019, 133, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Osadnik, T.; Bujak, K.; Osadnik, K.; Czarnecka, H.; Pawlas, N.; Reguła, R.; Fronczek, M.; Lejawa, M.; Gawlita, M.; Gonera, M.; et al. Novel inflammatory biomarkers may reflect subclinical inflammation in young healthy adults with obesity. Endokrynol. Pol. 2019, 70, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C.; Ahluwalia, N.; Albers, R.; Bosco, N.; Bourdet-Sicard, R.; Haller, D.; Holgate, S.T.; Jönsson, L.S.; Latulippe, M.E.; Marcos, A.; et al. A Consideration of Biomarkers to be Used for Evaluation of Inflammation in Human Nutritional Studies. Br. J. Nutr. 2013, 109, S1–S34. [Google Scholar] [CrossRef] [Green Version]
- Hickman, D.L. Evaluation of the neutrophil:lymphocyte ratio as an indicator of chronic distress in the laboratory mouse. Lab. Anim. (NY) 2017, 46, 303–307. [Google Scholar] [CrossRef] [Green Version]
- Swan, M.P.; Hickman, D.L. Evaluation of the neutrophil-lymphocyte ratio as a measure of distress in rats. Lab. Anim. (NY) 2014, 43, 276–282. [Google Scholar] [CrossRef]
- Afriandi, H.Y.; Gatot, P.A. Correlation of neutrophils lymphocytes ratio with femur muscle damage due to acute limb ischemia in white Wistar rats. Bali. Med. J. 2020, 9, 99–103. [Google Scholar] [CrossRef]
- Dolma, T.; Mukherjee, R.; Mukherjee, R.; Pati, B.K.; De, U.K. Acute Phase Response and Neutrophils: Lymphocyte Ratio in Response to Astaxanthin in Staphylococcal Mice Mastitis Model. J. Vet. Med. 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Marx, G. Fluid therapy in sepsis with capillary leakage. Eur. J. Anaesthesiol. 2003, 20, 429–442. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, K.E.; Mikkola, A.M.; Stepanek, A.M.; Vernet, A.; Hall, C.D.; Sun, C.C.; Yildirim, E.; Staropoli, J.F.; Lee, J.T.; Brown, D.E. Practical murine hematopathology: A comparative review and implications for research. Comp. Med. 2015, 65, 96–113, PMCID:PMC4408895. [Google Scholar] [PubMed]
- Qiu, J.; Xiaodi, N.; Wang, J.; Xing, Y.; Leng, B.; Dong, J.; Li, H.; Luo, M.; Zhang, Y.; Dai, X.; et al. Capsaicin Protects Mice from Community-Associated Methicillin-Resistant Staphylococcus aureus Pneumonia. PLoS ONE 2012, 7, e33032. [Google Scholar] [CrossRef] [Green Version]
- Bonaventura, G.; Pompilio, A.; Zappacosta, R.; Petrucci, F.; Fiscarelli, E.; Rossi, C.; Piccolomini, R. Role of Excessive Inflammatory Response to Stenotrophomonas maltophilia Lung Infection in DBA/2 Mice and Implications for Cystic Fibrosis. Infect. Imunn. 2010, 78, 2466–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushi, M.; Ito, T.; Oka, T.; Kitazawa, T.; Miyoshi-Akiyama, T.; Kirikae, T.; Yamashita, M.; Kudo, K. Serial Histopathological Examination of the Lungs of Mice Infected with Influenza A Virus PR8 Strain. PLoS ONE 2011, 6, e21207. [Google Scholar] [CrossRef] [Green Version]
- Fukushi, M.; Yamashita, M.; Miyoshi-Akiyama, T.; Kubo, S.; Yamamoto, K.; Kudo, K. Laninamivir Octanoate and Artificial Surfactant Combination Therapy Significantly Increases Survival of Mice Infected with Lethal Influenza H1N1 Virus. PLoS ONE 2012, 7, e42419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aeffner, F.; Bolon, B.; Davis, I.C. Mouse Models of Acute Respiratory Distress Syndrome: A Review of Analytical Approaches, Pathologic Features, and Common Measurements. Toxicol. Pathol. 2015, 43, 1074–1092. [Google Scholar] [CrossRef] [PubMed]
- Sordi, R.; Menezes-de-Lima, O.; Della-Justina, A.M.; Rezende, E.; Assreuy, J. Pneumonia-induced sepsis in mice: Temporal study of inflammatory and cardiovascular parameters. Int. J. Exp. Pathol. 2013, 94, 144–155. [Google Scholar] [CrossRef]
- Chastre, J.; Raghu, G.; Soler, P.; Brun, P.; Basset, F.; Gibert, C. Pulmonary Fibrosis following Pneumonia Due to Acute Legionnaires’ Disease: Clinical, Ultrastructural, and Immunofluorescent Study. Chest 1987, 91, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Carrington, C.B. Pathology of Legionnaires’ disease. Ann. Intern. Med. 1979, 90, 496–499. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trousil, J.; Frgelecová, L.; Kubíčková, P.; Řeháková, K.; Drašar, V.; Matějková, J.; Štěpánek, P.; Pavliš, O. Acute Pneumonia Caused by Clinically Isolated Legionella pneumophila Sg 1, ST 62: Host Responses and Pathologies in Mice. Microorganisms 2022, 10, 179. https://doi.org/10.3390/microorganisms10010179
Trousil J, Frgelecová L, Kubíčková P, Řeháková K, Drašar V, Matějková J, Štěpánek P, Pavliš O. Acute Pneumonia Caused by Clinically Isolated Legionella pneumophila Sg 1, ST 62: Host Responses and Pathologies in Mice. Microorganisms. 2022; 10(1):179. https://doi.org/10.3390/microorganisms10010179
Chicago/Turabian StyleTrousil, Jiří, Lucia Frgelecová, Pavla Kubíčková, Kristína Řeháková, Vladimír Drašar, Jana Matějková, Petr Štěpánek, and Oto Pavliš. 2022. "Acute Pneumonia Caused by Clinically Isolated Legionella pneumophila Sg 1, ST 62: Host Responses and Pathologies in Mice" Microorganisms 10, no. 1: 179. https://doi.org/10.3390/microorganisms10010179