Resistant Genes and Multidrug-Resistant Bacteria in Wastewater: A Study of Their Transfer to the Water Reservoir in the Czech Republic
Abstract
:1. Introduction
2. Materials and Methods
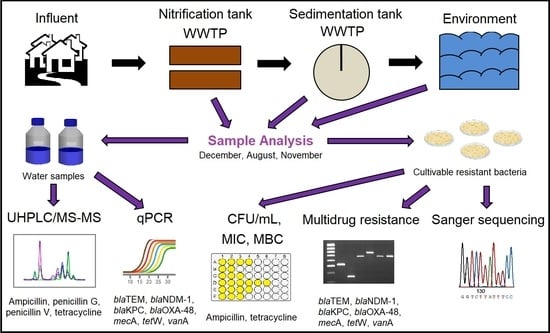
2.1. Wastewater and Environmental Sampling
2.2. Determination of Counts of Non-Resistant and Resistant Bacteria
2.3. Measurement of Growth Curves of Resistant Isolates
2.4. Determination of MIC and MBC Values
2.5. Antibiotic Determination
2.6. Isolation of Bacterial DNA from Water
2.7. Relative Abundance of ARGs in Water Samples
2.8. Evaluation of Multidrug Resistance of Bacterial Isolates
2.9. Identification of Resistant Bacteria
2.10. Statistical Analysis
3. Results and Discussion
3.1. Antibiotic Determination
3.2. Quantification of ARGs in Water
3.3. Characteristics of Ampicillin-Resistant Bacteria from Water Samples
3.4. Identification of Bacterial Isolates and Evaluation of Multidrug Resistance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Nina, P.B.; Jp, D.; Kumar, S.; Singh, B.; Tiwari, R.R. Futuristic Non-antibiotic Therapies to Combat Antibiotic Resistance: A Review. Front. Microbiol. 2021, 12, 609459. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, X.X.; Ye, L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE 2011, 6, e26041. [Google Scholar] [CrossRef] [Green Version]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Czekalski, N.; Sigdel, R.; Birtel, J.; Matthews, B.; Bürgmann, H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015, 81, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Suda, K.J.; Hicks, L.A.; Roberts, R.M.; Hunkler, R.J.; Taylor, T.H. Trends and seasonal variation in outpatient antibiotic prescription rates in the United States, 2006 to 2010. Antimicrob. Agents Chemother. 2014, 58, 2763–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Klein, E.Y.; Laxminarayan, R. Seasonality and temporal correlation between community antibiotic use and resistance in the United States. Clin. Infect Dis. 2012, 55, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makowska, N.; Bresa, K.; Koczura, R.; Philips, A.; Nowis, K.; Mokracka, J. Urban wastewater as a conduit for pathogenic Gram-positive bacteria and genes encoding resistance to β-lactams and glycopeptides. Sci. Total Environ. 2021, 765, 144176. [Google Scholar] [CrossRef]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Niestępski, S. Quantitative Occurrence of Antibiotic Resistance Genes among Bacterial Populations from Wastewater Treatment Plants Using Activated Sludge. Appl. Sci. 2019, 9, 387. [Google Scholar] [CrossRef] [Green Version]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Korzeniewska, E.; Harnisz, M. Relationship between modification of activated sludge wastewater treatment and changes in antibiotic resistance of bacteria. Sci. Total Environ. 2018, 639, 304–315. [Google Scholar] [CrossRef]
- Zieliński, W.; Buta, M.; Hubeny, J.; Korzeniewska, E.; Harnisz, M.; Nowrotek, M.; Płaza, G. Prevalence of Beta Lactamases Genes in Sewage and Sludge Treated in Mechanical-Biological Wastewater Treatment Plants. Ecol. Eng. 2019, 20, 80–86. [Google Scholar] [CrossRef]
- Mbanga, J.; Amoako, D.G.; Abia, A.L.K.; Allam, M.; Ismail, A.; Essack, S.Y. Genomic Insights of Multidrug-Resistant Escherichia coli From Wastewater Sources and Their Association With Clinical Pathogens in South Africa. Front. Vet. Sci. 2021, 8, 636715. [Google Scholar] [CrossRef]
- Lépesová, K.; Olejníková, P.; Mackuľak, T.; Cverenkárová, K.; Krahulcová, M.; Bírošová, L. Hospital Wastewater—Important Source of Multidrug Resistant Coliform Bacteria with ESBL-Production. Int. J. Environ. Res. Public Health 2020, 17, 7827. [Google Scholar] [CrossRef] [PubMed]
- Chia, P.; Sengupta, S.; Kukreja, A.; Ponnampalavanar, S.S.L.; Ng, O.T.; Marimuthu, K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob. Resist. Infect. Control 2020, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- Mody, L.; Washer, L.L.; Kaye, K.S.; Gibson, K.; Saint, S.; Reyes, K.; Cassone, M.; Mantey, J.; Cao, J.; Altamimi, S.; et al. Multidrug-resistant Organisms in Hospitals: What Is on Patient Hands and in Their Rooms? Clin. Infect Dis. 2019, 69, 1837–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, E.A.; Garzón, L.M.; Gómez, I.D.; Jiménez, J.N. Multidrug resistance and diversity of resistance profiles in carbapenem-resistant Gram-negative bacilli throughout a wastewater treatment plant in Colombia. J. Glob. Antimicrob. Resist. 2020, 22, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 2009, 276, 2521–2530. [Google Scholar] [CrossRef] [Green Version]
- Stachurová, T.; Piková, H.; Bartas, M.; Semerád, J.; Svobodová, K.; Malachová, K. Beta-lactam resistance development during the treatment processes of municipal wastewater treatment plants. Chemosphere 2021, 280, 130749. [Google Scholar] [CrossRef]
- Honda, R.; Tachi, C.; Yasuda, K.; Hirata, T.; Noguchi, M.; Hara-Yamamura, H.; Yamamoto-Ikemoto, R.; Watanabe, T. Estimated discharge of antibiotic-resistant bacteria from combined sewer overflows of urban sewage system. npj Clean Water 2020, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Barancheshme, F.; Munir, M. Strategies to combat antibiotic resistance in the wastewater plants. Front. Microbiol. 2018, 8, 2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SUKL (State Institution of Drug Control). Consumption of Antibiotics in the Czech Republic in the Years 2008–2018—Part 1. Available online: https://www.sukl.cz/file/92111_1_1 (accessed on 12 January 2022).
- SUKL (State Institution of Drug Control). Consumption of Antibiotics in the Czech Republic in the Years 2008–2018—Part 2. Available online: https://www.sukl.cz/file/92415_1_1 (accessed on 12 January 2022).
- Fouz, N.; Pangesti, K.N.A.; Yasir, M.; Al-Malki, A.L.; Azhar, E.I.; Hill-Cawthorne, G.A.; Abd El Ghany, M. The Contribution of Wastewater to the Transmission of Antimicrobial Resistance in the Environment: Implications of Mass Gathering Settings. Trop. Med. Infect. Dis. 2020, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Klančnik, A.; Piskernik, S.; Jeršek, B.; Smole Možina, S. Evaluation of diffusion and dilution methods to determine the antibacterial activity of plant extracts. J. Microbiol. Methods 2010, 81, 121–126. [Google Scholar] [CrossRef]
- Svobodová, K.; Semerád, J.; Petráčková, D.; Novotný, Č. Antibiotic resistance in Czech urban wastewater treatment plants: Microbial and molecular genetic characterization. Microb. Drug Resist. 2018, 24, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Stach, J.E.M.; Bathe, S.; Clapp, J.P.; Burns, R.G. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. Ecol. 2001, 36, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Mlynarcik, P.; Roderova, M.; Kolar, M. Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae. Jundishapur J. Microbiol. 2016, 9, e29314. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, P.C.S.; Monteiro, A.S.; Marques, S.G.; Monteiro, S.G.; Monteiro-Neto, V.; Coqueiro, M.M.; Marques, A.C.; de Jesus Gomes Turri, R.; Santos, S.G. Phenotypic and molecular detection of the blaKPC gene in clinical isolates from inpatients at hospitals in São Luis, MA, Brazil. BMC Infect. Dis. 2016, 16, 737. [Google Scholar] [CrossRef] [Green Version]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Rathnayake, I.U.; Hargreaves, M.; Huygens, F. Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates. Syst. Appl. Microbiol. 2012, 35, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novo, A.; Manaia, C. Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2010, 87, 1157–1166. [Google Scholar] [CrossRef]
- Mukherjee, M.; Laird, E.; Gentry, T.J.; Brooks, J.P.; Karthikeyan, R. Increased Antimicrobial and Multidrug Resistance Downstream of Wastewater Treatment Plants in an Urban Watershed. Front. Microbiol. 2021, 12, 657353. [Google Scholar] [CrossRef]
- Kulkarni, P.; Olson, N.D.; Raspanti, G.A.; Rosenberg Goldstein, R.E.; Gibbs, S.G.; Sapkota, A.; Sapkota, A.R. Antibiotic Concentrations Decrease during Wastewater Treatment but Persist at Low Levels in Reclaimed Water. Int. J. Environ. Res. Public Health 2017, 14, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulkowska, A.; Leung, H.W.; So, M.K.; Taniyasu, S.; Yamashita, N.; Yeung, L.W.Y.; Richardson, B.J.; Lei, A.P.; Giesy, J.P.; Lam, P.K.S. Removal of antibiotics from wastewater by sewage treatment facilities in Hong Kong and Shenzhen, China. Water Res. 2008, 42, 395–403. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Sui, Q.; Tong, J.; Jiang, C.; Lu, X.; Zhang, Y.; Wei, Y. Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res. 2016, 91, 339–349. [Google Scholar] [CrossRef]
- Hembach, N.; Alexander, J.; Hiller, C.; Wieland, A.; Schwartz, T. Dissemination prevention of antibiotic resistant and facultative pathogenic bacteria by ultrafiltration and ozone treatment at an urban wastewater treatment plant. Sci. Rep. 2019, 9, 12843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafraf, I.D.; Lekunberri, I.; Sànchez-Melsió, A.; Aouni, M.; Borrego, C.M.; Balcázar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, W.; Guo, G.; Dong, C.; Shen, Y.; Qin, S.; Chen, L.; Zhang, W. An Escherichia coli isolate from hospital sewage carries blaNDM-1 and blaoxa-10. Arch. Microbiol. 2021, 203, 4427–4432. [Google Scholar] [CrossRef]
- Ebomah, K.E.; Okoh, A.I. Enterobacter cloacae harbouring blaNDM-1, blaKPC, and blaOXA-48-like carbapenem-resistant genes isolated from different environmental sources in South Africa. Int. J. Environ. Sci. 2021, 78, 151–164. [Google Scholar] [CrossRef]
- Thakali, O.; Malla, B.; Tandukar, S.; Sthapit, N.; Raya, S.; Furukawa, T.; Sei, K.; Sherchand, J.B.; Haramoto, E. Release of Antibiotic-Resistance Genes from Hospitals and a Wastewater Treatment Plant in the Kathmandu Valley, Nepal. Water 2021, 13, 2733. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, X.; Huang, H.; Zhang, J. Prevalence of Antibiotic Resistance Genes and Their Association with Antibiotics in a Wastewater Treatment Plant: Process Distribution and Analysis. Water 2019, 11, 2495. [Google Scholar] [CrossRef] [Green Version]
- Oravcova, V.; Mihalcin, M.; Zakova, J.; Pospisilova, L.; Masarikova, M.; Literak, I. Vancomycin-resistant enterococci with vanA gene in treated municipal wastewater and their association with human hospital strains. Sci. Total Environ. 2017, 609, 633–643. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal infection-treatment and antibiotic resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 1–99. [Google Scholar]
- Caucci, S.; Karkman, A.; Cacace, D.; Rybicki, M.; Timpel, P.; Voolaid, V.; Gurke, R.; Virta, M.; Berendonk, T.U. Seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol. Ecol. 2016, 92, fiw060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obayiuwana, A.; Ogunjobi, A.; Yang, M.; Ibekwe, M. Characterization of Bacterial Communities and Their Antibiotic Resistance Profiles in Wastewaters Obtained from Pharmaceutical Facilities in Lagos and Ogun States, Nigeria. Int. J. Environ. Res. Public Health 2018, 15, 1365. [Google Scholar] [CrossRef] [Green Version]
- Aslan, A.; Cole, Z.; Bhattacharya, A.; Oyibo, O. Presence of Antibiotic-Resistant Escherichia coli in Wastewater Treatment Plant Effluents Utilized as Water Reuse for Irrigation. Water 2018, 10, 805. [Google Scholar] [CrossRef] [Green Version]
- Stachurová, T.; Malachová, K.; Semerád, J.; Sterniša, M.; Rybková, Z.; Možina, S.S. Tetracycline Induces the Formation of Biofilm of Bacteria from Different Phases of Wastewater Treatment. Processes 2020, 8, 989. [Google Scholar] [CrossRef]
- Harnisz, M.; Kiedrzyńska, E.; Kiedrzyński, M.; Korzeniewska, E.; Czatzkowska, M.; Koniuszewska, I.; Jóźwik, A.; Szklarek, S.; Niestępski, S.; Zalewski, M. The impact of WWTP size and sampling season on the prevalence of antibiotic resistance genes in wastewater and the river system. Sci. Total Environ. 2020, 741, 140466. [Google Scholar] [CrossRef]
- Skwor, T.; Stringer, S.; Haggerty, J.; Johnson, J.; Duhr, S.; Johnson, M.; Seckinger, M.; Stemme, M. Prevalence of Potentially Pathogenic Antibiotic-Resistant Aeromonas spp. in Treated Urban Wastewater Effluents versus Recipient Riverine Populations: A 3-Year Comparative Study. Appl. Environ. Microbiol. 2020, 86, e02053-19. [Google Scholar] [CrossRef]
- Oliveira, H.; Pinto, G.; Oliveira, A.; Noben, J.P.; Hendrix, H.; Lavigne, R.; Łobocka, M.; Kropinski, A.M.; Azeredo, J. Characterization and genomic analyses of two newly isolated Morganella phages define distant members among Tevenvirinae and Autographivirinae subfamilies. Sci. Rep. 2017, 7, 46157. [Google Scholar] [CrossRef] [Green Version]
- Luczkiewicz, A.; Kotlarska, E.; Artichowicz, W.; Tarasewicz, K.; Fudala-Ksiaze, S. Antimicrobial resistance of Pseudomonas spp. isolated from wastewater and wastewater-impacted marine coastal zone. Environ. Sci. Pollut. Res. 2015, 22, 19823–19834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowska, M.; Kowalska, S.; Popowska, M. Diversity of β-lactam resistance genes in gram-negative rods isolated from a municipal wastewater treatment plant. Ann. Microbiol. 2019, 69, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Alam, A.N.; Sarvari, J.; Motamedifar, M.; Khoshkharam, H.; Yousefi, M.; Moniri, R.; Bazargani, A. The occurrence of blaTEM, blaSHV and blaOXA genotypes in Extended-Spectrum β-Lactamase (ESBL)-producing Pseudomonas aeruginosa strains in Southwest of Iran. Gene Rep. 2018, 13, 19–23. [Google Scholar] [CrossRef]
- Mahrouki, S.; Perilli, M.; Bourouis, A.; Chihi, H.; Ferjani, M.; Moussa, M.B.; Amicosante, G.; Belhadj, O. Prevalence of quinolone resistance determinant qnrA6 among broad- and extended-spectrum beta-lactam-resistant Proteus mirabilis and Morganella morganii clinical isolates with sul1-type class 1 integron association in a Tunisian hospital. Scand. J. Infect. Dis. 2013, 45, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Obayiuwana, A.; Ibekwe, A.M. Antibiotic Resistance Genes Occurrence in Wastewaters from Selected Pharmaceutical Facilities in Nigeria. Water 2020, 12, 1897. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Devatha, C.P.; Pavithra, N. Isolation and identification of Pseudomonas from wastewater, its immobilization in cellulose biopolymer and performance in degrading Triclosan. J. Environ. Manag. 2019, 232, 584–591. [Google Scholar] [CrossRef]
- Batrich, M.; Maskeri, L.; Schubert, R.; Ho, B.; Kohout, M.; Abdeljaber, M.; Abuhasna, A.; Kholoki, M.; Psihogios, P.; Razzaq, T.; et al. Pseudomonas Diversity Within Urban Freshwaters. Front. Microbiol. 2019, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Etayo, L.; González, D.; Leiva, J.; Vitas, A.I. Multidrug-Resistant Bacteria Isolated from Different Aquatic Environments in the North of Spain and South of France. Microorganisms 2020, 8, 1425. [Google Scholar] [CrossRef] [PubMed]
- Amador, P.P.; Fernandes, R.M.; Prudêncio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of Class A and Class C β-lactamases. J. Environ. Sci. Health A 2015, 50, 26–39. [Google Scholar] [CrossRef]





| Sampling | Water Sample | Without AMP | 500 mg/L AMP |
|---|---|---|---|
| log CFU/mL | |||
| A | Nitrification tank WWTP | 6.410 ± 0.067 Abc | 6.006 ± 0.015 Abc |
| Sedimentation tank WWTP | 3.452 ± 0.054 a | 2.694 ± 0.013 * a | |
| Dam | 3.034 ± 0.080 a | 2.041 ± 0.001 * a | |
| B | Nitrification tank WWTP | 6.300 ± 0.022 bc | 5.581 ± 0.062 * bc |
| Sedimentation tank WWTP | 4.019 ± 0.023 a | 3.423 ± 0.025 * a | |
| Dam | 3.349 ± 0.048 a | 2.292 ± 0.088 * a | |
| C | Nitrification tank WWTP | 6.067 ± 0.164 bc | 5.818 ± 0.195 bc |
| Sedimentation tank WWTP | 3.554 ± 0.239 a | 2.970 ± 0.099 a | |
| Dam | 2.540 ± 0.062 a | 1.739 ± 0.040 * a | |
| Isolate | blaTEM | blaNDM-1 | tetW | vanA | Identification | Accession Number |
|---|---|---|---|---|---|---|
| N1_A | + | + | + | - | Aeromonas sp. | OL832061 |
| N2_A | + | - | + | - | Aeromonas sp. | OL832062 |
| N3_A | + | - | + | - | Aeromonas sp. | OL832063 |
| N4_A | + | + | + | - | Aeromonas sp. | OL832064 |
| S1_A | + | - | + | - | Aeromonas sp. | OL832065 |
| S2_A | + | + | + | - | Aeromonas sp. | OL832066 |
| S3_A | + | - | - | - | Aeromonas sp. | OL832067 |
| D1_A | + | - | + | + | Aeromonas sp. | OL832068 |
| N1_B | + | - | + | - | Aeromonas sp. | OL832069 |
| N2_B | + | - | + | - | Aeromonas sp. | OL832070 |
| N3_B | + | - | + | - | Aeromonas sp. | OL832071 |
| N4_B | + | - | + | - | Aeromonas sp. | OL832072 |
| N5_B | + | - | + | - | Aeromonas sp. | OL832073 |
| N6_B | + | - | + | - | Aeromonas sp. | OL832074 |
| N7_B | + | - | + | - | Aeromonas sp. | OL832075 |
| N8_B | + | + | - | - | Aeromonas sp. | OL832076 |
| N9_B | + | - | + | - | Aeromonas sp. | OL832077 |
| N10_B | + | - | + | - | Aeromonas sp. | OL832078 |
| S1_B | + | - | + | - | Aeromonas sp. | OL832079 |
| S2_B | + | - | + | - | Aeromonas sp. | OL832080 |
| S3_B | + | - | - | - | Aeromonas sp. | OL832081 |
| S4_B | + | - | + | - | Morganella sp. | OL832082 |
| S5_B | + | - | + | - | Aeromonas sp. | OL832083 |
| S6_B | + | - | + | - | Aeromonas sp. | OL832084 |
| S7_B | + | - | + | - | Aeromonas sp. | OL832085 |
| S8_B | + | - | + | - | Aeromonas sp. | OL832086 |
| S9_B | + | + | + | - | Aeromonas sp. | OL832087 |
| S10_B | + | - | + | - | Morganella sp. | OL832088 |
| D1_B | + | - | + | - | Aeromonas sp. | OL832089 |
| D2_B | + | - | - | - | Aeromonas sp. | OL832090 |
| D3_B | + | - | + | - | Aeromonas sp. | OL832091 |
| D4_B | + | - | + | - | Pseudomonas sp. | OL832092 |
| D5_B | + | - | + | - | Aeromonas sp. | OL832093 |
| D6_B | + | + | + | - | Pseudomonas sp. | OL832094 |
| D7_B | + | + | + | - | Pseudomonas sp. | OL832095 |
| D8_B | + | - | - | - | Aeromonas sp. | OL832096 |
| D9_B | + | - | - | - | Aeromonas sp. | OL832097 |
| D10_B | + | - | + | - | Aeromonas sp. | OL832098 |
| N1_C | + | - | + | - | Aeromonas sp. | OL832099 |
| N2_C | + | + | + | - | Aeromonas sp. | OL832100 |
| N3_C | + | - | + | - | Aeromonas sp. | OL832101 |
| N4_C | + | - | + | - | Aeromonas sp. | OL832102 |
| N5_C | + | - | + | - | Aeromonas sp. | OL832103 |
| S1_C | + | - | + | - | Aeromonas sp. | OL832104 |
| S2_C | + | - | - | + | Aeromonas sp. | OL832105 |
| S3_C | + | + | + | - | Aeromonas sp. | OL832106 |
| S4_C | + | - | - | - | Aeromonas sp. | OL832107 |
| S5_C | + | - | + | - | Aeromonas sp. | OL832108 |
| D1_C | + | - | + | + | Aeromonas sp. | OL832109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stachurová, T.; Sýkorová, N.; Semerád, J.; Malachová, K. Resistant Genes and Multidrug-Resistant Bacteria in Wastewater: A Study of Their Transfer to the Water Reservoir in the Czech Republic. Life 2022, 12, 147. https://doi.org/10.3390/life12020147
Stachurová T, Sýkorová N, Semerád J, Malachová K. Resistant Genes and Multidrug-Resistant Bacteria in Wastewater: A Study of Their Transfer to the Water Reservoir in the Czech Republic. Life. 2022; 12(2):147. https://doi.org/10.3390/life12020147
Chicago/Turabian StyleStachurová, Tereza, Nikola Sýkorová, Jaroslav Semerád, and Kateřina Malachová. 2022. "Resistant Genes and Multidrug-Resistant Bacteria in Wastewater: A Study of Their Transfer to the Water Reservoir in the Czech Republic" Life 12, no. 2: 147. https://doi.org/10.3390/life12020147