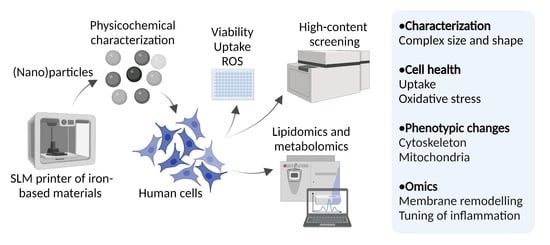
A Novel Nanosafety Approach Using Cell Painting, Metabolomics, and Lipidomics Captures the Cellular and Molecular Phenotypes Induced by the Unintentionally Formed Metal-Based (Nano)Particles
Abstract
:1. Introduction
2. Results and Discussion
2.1. Physicochemical Characterization, the Metal Release, and the Endotoxin Levels

2.2. Cell Viability, Oxidative Stress, and the (Nano)Particle Internalization

2.3. Profiling of the Cell’s Morphological Phenotypes by the Cell Painting Assay


2.4. Biological Implications of the Cell Painting Features
2.5. Curation Strategies for the Cell Painting Datasets
2.6. Lipidomics

2.7. Targeted Metabolomics
3. Conclusions
- A novel method for the nanosafety studies is described and employed as capable of detecting the early changes in the cell morphological phenotypes at low (nano)particle concentrations and able to suggest the prevailing adverse MoAs induced by the (nano)particle-cell interactions. This indicates that the cell stress conditions may be detected upon exposure to the (nano)particles before it can be observed in the reduced cell viability;
- The initial integration of the techniques provides important knowledge for the morphological, lipidomic, and metabolomic signatures as biomarkers of the AMP exposure;
- A proof-of-concept is presented that suggests that the MoAs of the AMPs are complex and, especially at the molecular level, do not always follow a concentration-dependent pattern. We envision future studies to comprehensively elucidate the AMP-cell interactions and MoA in human cells, and to apply lung/bronchial epithelial cells and macrophages as cell models in the Cell Painting profiling.
4. Materials and Methods
4.1. AMPs and the Characterization Methods
4.1.1. Source, Stock Dispersions, and Endotoxin Test
4.1.2. Scanning Electron Microscopy Combined with Energy Dispersive Spectroscopy (SEM-EDS)
4.1.3. Transmission Electron Microscopy (TEM)
4.1.4. X-ray Photoelectron Spectroscopy (XPS)
4.1.5. Analysis of the Metal Release in the Cell Medium by Atomic Absorption Spectroscopy (AAS)
4.2. Mono- and Co-Culture Cell Models
4.3. Cell Viability and Reactive Oxygen Species (ROS) Detection Assays
4.4. AMPs Internalization Analysis
4.5. Cell Painting and Data Analysis
4.5.1. Cell Seeding and AMP Exposure
4.5.2. Cell Staining and Image Acquisition
4.5.3. Image Processing and Cell Profiling
4.5.4. Univariate, Unsupervised, and Supervised Multivariate Analyses
4.6. Multiplex Immunoassay
4.7. Lipidomic Analysis
4.8. Metabolomic Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wong, K.V.; Hernandez, A. A review of additive manufacturing. Int. Sch. Res. Not. 2012, 2012, 208760. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.P.; Elangovan, S.; Mohanraj, R.; Ramakrishna, J.R. Review on the evolution and technology of State-of-the-Art metal additive manufacturing processes. Mater. Today Proc. 2021, 46, 7907–7920. [Google Scholar] [CrossRef]
- Alijagic, A.; Scherbak, N.; Kotlyar, O.; Karlsson, P.; Persson, A.; Hedbrant, A.; Norinder, U.; Larsson, M.; Felth, J.; Andersson, L.; et al. Cell Painting unveils cell response signatures to (nano) particles formed in additive manufacturing. Toxicol. Lett. 2022, 368, S226–S227. [Google Scholar] [CrossRef]
- Alijagic, A.; Engwall, M.; Särndahl, E.; Karlsson, H.; Hedbrant, A.; Andersson, L.; Karlsson, P.; Dalemo, M.; Scherbak, N.; Färnlund, K.; et al. Particle safety assessment in additive manufacturing: From exposure risks to advanced toxicology testing. Front. Toxicol. 2022, 4, 836447. [Google Scholar] [CrossRef] [PubMed]
- Runström Eden, G.; Tinnerberg, H.; Rosell, L.; Möller, R.; Almstrand, A.C.; Bredberg, A. Exploring Methods for Surveillance of Occupational Exposure from Additive Manufacturing in Four Different Industrial Facilities. Ann. Work Expo. Health 2022, 66, 163–177. [Google Scholar] [CrossRef]
- Vallabani, N.S.; Alijagic, A.; Persson, A.; Odnevall, I.; Särndahl, E.; Karlsson, H.L. Toxicity evaluation of particles formed during 3D-printing: Cytotoxic, genotoxic, and inflammatory response in lung and macrophage models. Toxicology 2022, 467, 153100. [Google Scholar] [CrossRef]
- Hartung, T.; FitzGerald, R.E.; Jennings, P.; Mirams, G.R.; Peitsch, M.C.; Rostami-Hodjegan, A.; Shah, I.; Wilks, M.F.; Sturla, S.J. Systems toxicology: Real world applications and opportunities. Chem. Res. Toxicol. 2017, 3, 870–882. [Google Scholar] [CrossRef]
- Fröhlich, E. Role of omics techniques in the toxicity testing of nanoparticles. J. Nanobiotechnol. 2017, 15, 84. [Google Scholar] [CrossRef]
- Nymark, P.; Bakker, M.; Dekkers, S.; Franken, R.; Fransman, W.; García-Bilbao, A.; Greco, D.; Gulumian, M.; Hadrup, N.; Halappanavar, S.; et al. Toward rigorous materials production: New approach methodologies have extensive potential to improve current safety assessment practices. Small 2020, 16, 1904749. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Bray, M.A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; Carpenter, A.E. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016, 11, 1757–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakal, C.; Aach, J.; Church, G.; Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 2007, 316, 1753–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Severin, Y.; Hale, B.D.; Mena, J.; Goslings, D.; Frey, B.M.; Snijder, B. Multiplexed high-throughput immune cell imaging reveals molecular health-associated phenotypes. Sci. Adv. 2022, 8, 5631. [Google Scholar] [CrossRef] [PubMed]
- Schneidewind, T.; Brause, A.; Pahl, A.; Burhop, A.; Mejuch, T.; Sievers, S.; Waldmann, H.; Ziegler, S. Morphological profiling identifies a common mode of action for small molecules with different targets. ChemBioChem 2020, 21, 3197–3207. [Google Scholar] [CrossRef]
- Nyffeler, J.; Willis, C.; Lougee, R.; Richard, A.; Paul-Friedman, K.; Harrill, J.A. Bioactivity screening of environmental chemicals using imaging-based high-throughput phenotypic profiling. Toxicol. Appl. Pharmacol. 2020, 389, 114876. [Google Scholar] [CrossRef]
- Akbarzadeh, M.; Deipenwisch, I.; Schoelermann, B.; Pahl, A.; Sievers, S.; Ziegler, S.; Waldmann, H. Morphological profiling by means of the Cell Painting assay enables identification of tubulin-targeting compounds. Cell Chem. Biol. 2021, 29, 1053–1064. [Google Scholar] [CrossRef]
- Schiff, L.; Migliori, B.; Chen, Y.; Carter, D.; Bonilla, C.; Hall, J.; Fan, M.; Tam, E.; Ahadi, S.; Fischbacher, B.; et al. Integrating deep learning and unbiased automated high-content screening to identify complex disease signatures in human fibroblasts. Nat. Commun. 2022, 13, 1590. [Google Scholar] [CrossRef] [PubMed]
- Way, G.P.; Kost-Alimova, M.; Shibue, T.; Harrington, W.F.; Gill, S.; Piccioni, F.; Becker, T.; Shafqat-Abbasi, H.; Hahn, W.C.; Carpenter, A.E.; et al. Predicting cell health phenotypes using image-based morphology profiling. Mol. Biol. Cell 2021, 32, 995–1005. [Google Scholar] [CrossRef]
- Pahl, A.; Sievers, S. The Cell Painting assay as a screening tool for the discovery of bioactivities in new chemical matter. In Systems Chemical Biology; Ziegler, S., Waldmann, H., Eds.; Humana Press: New York, NY, USA, 2019; pp. 115–126. [Google Scholar] [CrossRef]
- Rietdijk, J.; Tampere, M.; Pettke, A.; Georgiev, P.; Lapins, M.; Warpman-Berglund, U.; Spjuth, O.; Puumalainen, M.R.; Carreras-Puigvert, J. A phenomics approach for antiviral drug discovery. BMC Biol. 2021, 19, 156. [Google Scholar] [CrossRef]
- Rietdijk, J.; Aggarwal, T.; Georgieva, P.; Lapins, M.; Carreras-Puigvert, J.; Spjuth, O. Morphological profiling of environmental chemicals enables efficient and untargeted exploration of combination effects. Sci. Tot. Environ. 2022, 832, 155058. [Google Scholar] [CrossRef]
- Nassiri, I.; McCall, M.N. Systematic exploration of cell morphological phenotypes associated with a transcriptomic query. Nucleic Acids Res. 2018, 46, e116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, T.; Daneshian, M.; Kamp, H.; Bois, F.Y.; Clench, M.R.; Coen, M.; Donley, B.; Fischer, S.M.; Ekman, D.R.; Fabian, E.; et al. Metabolomics in toxicology and preclinical research. ALTEX 2013, 30, 209–225. [Google Scholar] [CrossRef] [Green Version]
- Ventura, C.; Torres, V.; Vieira, L.; Gomes, B.; Rodrigues, A.S.; Rueff, J.; Penque, D.; Silva, M.J. New “Omics” Approaches as Tools to Explore Mechanistic Nanotoxicology. In Nanotoxicology in Safety Assessment of Nanomaterials. Advances in Experimental Medicine and Biology; Louro, H., Silva, M.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2022; Volume 1357, pp. 179–194. [Google Scholar] [CrossRef]
- Halappanavar, S.; Van Den Brule, S.; Nymark, P.; Gaté, L.; Seidel, C.; Valentino, S.; Zhernovkov, V.; Høgh Danielsen, P.; De Vizcaya, A.; Wolff, H.; et al. Adverse outcome pathways as a tool for the design of testing strategies to support the safety assessment of emerging advanced materials at the nanoscale. Part. Fibre Toxicol. 2020, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Demokritou, P.; Kavouras, I.G.; Ferguson, S.T.; Koutrakis, P. Development of a high volume cascade impactor for toxicological and chemical characterization studies. Aerosol Sci. Technol. 2022, 36, 925–933. [Google Scholar] [CrossRef]
- Gatoo, M.A.; Naseem, S.; Arfat, M.Y.; Mahmood Dar, A.; Qasim, K.; Zubair, S. Physicochemical properties of nanomaterials: Implication in associated toxic manifestations. BioMed Res. Int. 2014, 2014, 498420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarrick, S.; Wei, Z.; Moelijker, N.; Derr, R.; Persson, K.A.; Hendriks, G.; Odnevall Wallinder, I.; Hedberg, Y.; Karlsson, H.L. High variability in toxicity of welding fume nanoparticles from stainless steel in lung cells and reporter cell lines: The role of particle reactivity and solubility. Nanotoxicol. 2019, 13, 1293–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gliga, A.R.; Skoglund, S.; Odnevall Wallinder, I.; Fadeel, B.; Karlsson, H.L. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: The role of cellular uptake, agglomeration and Ag release. Part. Fibre Toxicol. 2014, 11, 11. [Google Scholar] [CrossRef] [Green Version]
- Rahmati, M.; Silva, E.A.; Reseland, J.E.; Heyward, C.A.; Haugen, H.J. Biological responses to physicochemical properties of biomaterial surface. Chem. Soc. Rev. 2020, 49, 5178–5224. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Mai, S.; Wang, D.; Song, C. Investigation into spatter behavior during selective laser melting of AISI 316L stainless steel powder. Mater. Des. 2015, 87, 797–806. [Google Scholar] [CrossRef]
- Simonelli, M.; Tuck, C.; Aboulkhair, N.T.; Maskery, I.; Ashcroft, I.; Wildman, R.D.; Hague, R. A study on the laser spatter and the oxidation reactions during selective laser melting of 316L stainless steel, Al-Si10-Mg, and Ti-6Al-4V. Metall. Mater. Trans. 2015, 46, 3842–3851. [Google Scholar] [CrossRef]
- Ly, S.; Rubenchik, A.M.; Khairallah, S.A.; Guss, G.; Matthews, M.J. Metal vapor micro-jet controls material redistribution in laser powder bed fusion additive manufacturing. Sci. Rep. 2017, 7, 4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, A.T.; Kriewall, C.S.; Leu, M.C.; Newkirk, J.W.; Brown, B. Characterization of laser spatter and condensate generated during the selective laser melting of 304L stainless steel powder. Addit. Manuf. 2020, 31, 100904. [Google Scholar] [CrossRef]
- Hedberg, Y.S.; Wei, Z.; McCarrick, S.; Romanovski, V.; Theodore, J.; Westin, E.M.; Wagner, R.; Persson, K.A.; Karlsson, H.L.; Odnevall Wallinder, I. Welding fume nanoparticles from solid and flux-cored wires: Solubility, toxicity, and role of fluorides. J. Haz. Mat. 2021, 413, 125273. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, H.; Schmittner, M.; Duschl, A.; Horejs-Hoeck, J. Residual endotoxin contaminations in recombinant proteins are sufficient to activate human CD1c+ dendritic cells. PloS ONE 2014, 9, e113840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oostingh, G.J.; Casals, E.; Italiani, P.; Colognato, R.; Stritzinger, R.; Ponti, J.; Pfaller, T.; Kohl, Y.; Ooms, D.; Favilli, F.; et al. Problems and challenges in the development and validation of human cell-based assays to determine nanoparticle-induced immunomodulatory effects. Part. Fibre Toxicol. 2011, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Boraschi, D. Endotoxin contamination: A key element in the interpretation of nanosafety studies. Nanomedicine 2016, 11, 269–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Italiani, P.; Casals, E.; Tran, N.; Puntes, V.F.; Boraschi, D. Optimising the use of commercial LAL assays for the analysis of endotoxin contamination in metal colloids and metal oxide nanoparticles. Nanotoxicology 2015, 9, 462–473. [Google Scholar] [CrossRef] [Green Version]
- Longhin, E.M.; El Yamani, N.; Rundén-Pran, E.; Dusinska, M. The alamar blue assay in the context of safety testing of nanomaterials. Front. Toxicol. 2022, 4, 981701. [Google Scholar] [CrossRef]
- Paunovic, J.; Vucevic, D.; Radosavljevic, T.; Mandić-Rajčević, S.; Pantic, I. Iron-based nanoparticles and their potential toxicity: Focus on oxidative stress and apoptosis. Chem. Biol. Interact. 2020, 316, 108935. [Google Scholar] [CrossRef]
- Cappellini, F.; Hedberg, Y.; McCarrick, S.; Hedberg, J.; Derr, R.; Hendriks, G.; Odnevall Wallinder, I.; Karlsson, H.L. Mechanistic insight into reactivity and (geno) toxicity of well-characterized nanoparticles of cobalt metal and oxides. Nanotoxicology 2018, 12, 602–620. [Google Scholar] [CrossRef]
- Latvala, S.; Hedberg, J.; Di Bucchianico, S.; Möller, L.; Odnevall Wallinder, I.; Elihn, K.; Karlsson, H.L. Nickel release, ROS generation and toxicity of Ni and NiO micro-and nanoparticles. PloS ONE 2016, 11, e0159684. [Google Scholar] [CrossRef]
- Nel, A.; Xia, T.; Madler, L.; Li, N. Toxic potential of materials at the nanolevel. Science 2006, 311, 622–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendriks, G.; Derr, R.S.; Misovic, B.; Morolli, B.; Calleja, F.M.; Vrieling, H. The extended ToxTracker assay discriminates between induction of DNA damage, oxidative stress, and protein misfolding. Toxicol. Sci. 2016, 150, 190–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendriks, G.; Atallah, M.; Morolli, B.; Calléja, F.; Ras-Verloop, N.; Huijskens, I.; Raamsman, M.; van de Water, B.; Vrieling, H. The ToxTracker assay: Novel GFP reporter systems that provide mechanistic insight into the genotoxic properties of chemicals. Toxicol. Sci. 2012, 125, 285–298. [Google Scholar] [CrossRef] [PubMed]
- McCarrick, S.; Cappellini, F.; Kessler, A.; Moelijker, N.; Derr, R.; Hedberg, J.; Wold, S.; Blomberg, E.; Odnevall Wallinder, I.; Hendriks, G.; et al. ToxTracker reporter cell lines as a tool for mechanism-based (geno) toxicity screening of nanoparticles—Metals, oxides and quantum dots. Nanomaterials 2020, 10, 110. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, J.; Hao, L.; Bijli, K.M.; Chandler, J.D.; Orr, M.; Hu, X.; Jones, D.P.; Go, Y.M. From the cover: Manganese stimulates mitochondrial H2O2 production in SH-SY5Y human neuroblastoma cells over physiologic as well as toxicologic range. Toxicol. Sci. 2017, 155, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.R.; Fernandes, J.; Go, Y.M.; Jones, D.P. Redox dynamics of manganese as a mitochondrial life-death switch. Biochem. Biophys. Res. Comm. 2017, 482, 388–398. [Google Scholar] [CrossRef] [Green Version]
- Pfalzer, A.C.; Bowman, A.B. Relationships between essential manganese biology and manganese toxicity in neurological disease. Curr. Environ. Health Rep. 2017, 4, 223–228. [Google Scholar] [CrossRef]
- Yang, Y.; Bazhin, A.V.; Werner, J.; Karakhanova, S. Reactive oxygen species in the immune system. Int. Rev. Immunol. 2013, 32, 249–270. [Google Scholar] [CrossRef]
- Kroll, A.; Pillukat, M.H.; Hahn, D.; Schnekenburger, J. Interference of engineered nanoparticles with in vitro toxicity assays. Arch. Toxicol. 2012, 86, 1123–1136. [Google Scholar] [CrossRef]
- Zhao, J.; Riediker, M. Detecting the oxidative reactivity of nanoparticles: A new protocol for reducing artifacts. J. Nanopart. Res. 2014, 16, 2493. [Google Scholar] [CrossRef] [Green Version]
- Pandey, N.K.; Xiong, W.; Wang, L.; Chen, W.; Bui, B.; Yang, J.; Amador, E.; Chen, M.; Xing, C.; Athavale, A.A.; et al. Aggregation-induced emission luminogens for highly effective microwave dynamic therapy. Bioact. Mater. 2022, 7, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Shevchuk, A.; Ruenraroengsak, P.; Miragoli, M.; Thorley, A.J.; Klenerman, D.; Lab, M.J.; Tetley, T.D.; Gorelik, J.; Korchev, Y.E. Imaging single nanoparticle interactions with human lung cells using fast ion conductance microscopy. Nano Lett. 2014, 14, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafsdottir, S.M.; Ljosa, V.; Sokolnicki, K.L.; Anthony Wilson, J.; Walpita, D.; Kemp, M.M.; Petri Seiler, K.; Carrel, H.A.; Golub, T.R.; Schreiber, S.L.; et al. Multiplex cytological profiling assay to measure diverse cellular states. PloS ONE 2013, 8, e80999. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, J.C.; Arevalo, J.; Piccioni, F.; Bray, M.A.; Hartland, C.L.; Wu, X.; Brooks, A.N.; Berger, A.H.; Boehm, J.S.; Carpenter, A.E.; et al. Cell Painting predicts impact of lung cancer variants. Mol. Biol. Cell 2022, 33, mbc-E21. [Google Scholar] [CrossRef]
- de Almeida, M.S.; Susnik, E.; Drasler, B.; Taladriz-Blanco, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem. Soc. Rev. 2021, 50, 5397–5434. [Google Scholar] [CrossRef] [PubMed]
- Buyukhatipoglu, K.; Clyne, A.M. Superparamagnetic iron oxide nanoparticles change endothelial cell morphology and mechanics via reactive oxygen species formation. J. Biomed. Mater. Res. A 2011, 96, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Ispanixtlahuatl-Meráz, O.; Schins, R.P.; Chirino, Y.I. Cell type specific cytoskeleton disruption induced by engineered nanoparticles. Environ. Sci. Nano 2018, 5, 228–245. [Google Scholar] [CrossRef]
- Déciga-Alcaraz, A.; Delgado-Buenrostro, N.L.; Ispanixtlahuatl-Meráz, O.; Freyre-Fonseca, V.; Flores-Flores, J.O.; Ganem-Rondero, A.; Vaca-Paniagua, F.; del Pilar Ramos-Godinez, M.; Morales-Barcenas, R.; Sánchez-Pérez, Y.; et al. Irreversible disruption of the cytoskeleton as induced by non-cytotoxic exposure to titanium dioxide nanoparticles in lung epithelial cells. Chem. Biol. Interact. 2020, 323, 109063. [Google Scholar] [CrossRef]
- Wulf, E.; Deboben, A.; Bautz, F.A.; Faulstich, H.; Wieland, T. Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc. Natl. Acad. Sci. USA 1979, 76, 4498–4502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhang, H.; Liang, X.; Zhang, J.; Tao, W.; Zhu, X.; Chang, D.; Zeng, X.; Liu, G.; Mei, L. Iron oxide nanoparticles induce autophagosome accumulation through multiple mechanisms: Lysosome impairment, mitochondrial damage, and ER stress. Mol. Pharm. 2016, 13, 2578–2587. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.I.; Jou, M.J. Mitochondrial swelling and generation of reactive oxygen species induced by photoirradiation are heterogeneously distributed. In Mitochondrial Pathogenesis; Lee, H.K., DiMauro, S., Tanaka, M., Wei, Y.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 112–122. [Google Scholar] [CrossRef]
- Li, D.W.; He, H.; Lin, B.B.; Xu, Z.Q.; Jiang, F.L.; Liu, Y. Studies on the isolated mitochondrial damage induced by α-tocopheryl succinate and its interactions with human serum albumin. RSC Adv. 2014, 4, 3913–3919. [Google Scholar] [CrossRef]
- Barkhade, T.; Mahapatra, S.K.; Banerjee, I. Study of mitochondrial swelling, membrane fluidity and ROS production induced by nano-TiO2 and prevented by Fe incorporation. Toxicol. Res. 2019, 8, 711–722. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Ahlinder, L.; Ekstrand-Hammarström, B.; Geladi, P.; Österlund, L. Large uptake of titania and iron oxide nanoparticles in the nucleus of lung epithelial cells as measured by Raman imaging and multivariate classification. Biophys. J. 2013, 105, 310–319. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, A.V.; Margreiter, R.; Ausserlechner, M.J.; Hagenbuchner, J. The Complex Interplay between Mitochondria, ROS and Entire Cellular Metabolism. Antioxidants 2022, 11, 1995. [Google Scholar] [CrossRef]
- Seal, S.; Carreras-Puigvert, J.; Trapotsi, M.A.; Yang, H.; Spjuth, O.; Bender, A. Integrating cell morphology with gene expression and chemical structure to aid mitochondrial toxicity detection. bioRxiv 2022. [Google Scholar] [CrossRef]
- Stern, S.T.; Adiseshaiah, P.P.; Crist, R.M. Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity. Part. Fibre Toxicol. 2012, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Moghadam, B.Y.; Hou, W.C.; Corredor, C.; Westerhoff, P.; Posner, J.D. Role of nanoparticle surface functionality in the disruption of model cell membranes. Langmuir 2012, 28, 16318–16326. [Google Scholar] [CrossRef]
- Wei, X.; Jiang, W.; Yu, J.; Ding, L.; Hu, J.; Jiang, G. Effects of SiO2 nanoparticles on phospholipid membrane integrity and fluidity. J. Haz. Mat. 2015, 287, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, W.E.; Chang, H.H.; Dan, A.; Kenis, P.J.; Murphy, C.J.; Leckband, D.E. Gold nanoparticles disrupt actin organization and pulmonary endothelial barriers. Sci. Rep. 2020, 10, 13320. [Google Scholar] [CrossRef] [PubMed]
- Bereiter-Hahn, J.; Vöth, M. Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994, 27, 198–219. [Google Scholar] [CrossRef] [PubMed]
- Fenton, A.R.; Jongens, T.A.; Holzbaur, E.L. Mitochondrial dynamics: Shaping and remodeling an organelle network. Curr. Opin. Cell Biol. 2021, 68, 28–36. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv 2018. [Google Scholar] [CrossRef]
- Kensert, A.; Harrison, P.J.; Spjuth, O. Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS Discov. 2019, 24, 466–475. [Google Scholar] [CrossRef] [Green Version]
- Seal, S.; Yang, H.; Vollmers, L.; Bender, A. Comparison of cellular morphological descriptors and molecular fingerprints for the prediction of cytotoxicity-and proliferation-related assays. Chem. Res. Toxicol. 2021, 34, 422–437. [Google Scholar] [CrossRef]
- Yeung, T.; Ozdamar, B.; Paroutis, P.; Grinstein, S. Lipid metabolism and dynamics during phagocytosis. Curr. Opin. Cell Biol. 2006, 18, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Sancak, Y.; Frasor, J.; Atilla-Gokcumen, G.E. A protective role for triacylglycerols during apoptosis. Biochemistry 2018, 57, 72–80. [Google Scholar] [CrossRef]
- Soria, N.C.; Aga, D.S.; Atilla-Gokcumen, G.E. Lipidomics reveals insights on the biological effects of copper oxide nanoparticles in a human colon carcinoma cell line. Mol. Omics 2019, 15, 30–38. [Google Scholar] [CrossRef]
- Chiurchiù, V.; Leuti, A.; Maccarrone, M. Bioactive lipids and chronic inflammation: Managing the fire within. Front. Immunol. 2018, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Vogel, A.; Brunner, J.S.; Hajto, A.; Sharif, O.; Schabbauer, G. Lipid scavenging macrophages and inflammation. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2022, 1867, 159066. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, X.; Liu, X.; Zheng, J. Co-culture of human alveolar epithelial (A549) and macrophage (THP-1) cells to study the potential toxicity of ambient PM2. 5: A comparison of growth under ALI and submerged conditions. Toxicol. Res. 2020, 9, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Li, X.B.; Zheng, H.; Zhang, Z.R.; Li, M.; Huang, Z.Y.; Schluesener, H.J.; Li, Y.Y.; Xu, S.Q. Glia activation induced by peripheral administration of aluminum oxide nanoparticles in rat brains. Nanomed. Nanotechnol. Biol. Med. 2009, 5, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, K.; Davoren, M.; Boertz, J.; Schins, R.P.; Hoffmann, E.; Dopp, E. Titanium dioxide nanoparticles induce oxidative stress and DNA-adduct formation but not DNA-breakage in human lung cells. Part. Fibre Toxicol. 2009, 6, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barelli, H.; Antonny, B. Lipid unsaturation and organelle dynamics. Curr. Opin. Cell Biol. 2016, 41, 25–32. [Google Scholar] [CrossRef]
- Himly, M.; Geppert, M.; Hofer, S.; Hofstätter, N.; Horejs-Höck, J.; Duschl, A. When would immunologists consider a nanomaterial to be safe? Recommendations for planning studies on nanosafety. Small 2020, 16, 1907483. [Google Scholar] [CrossRef]
- Boraschi, D.; Alijagic, A.; Auguste, M.; Barbero, F.; Ferrari, E.; Hernadi, S.; Mayall, C.; Michelini, S.; Navarro Pacheco, N.I.; Prinelli, A.; et al. Addressing nanomaterial immunosafety by evaluating innate immunity across living species. Small 2020, 16, 2000598. [Google Scholar] [CrossRef]
- Zhang, Q.; Guo, X.; Cheng, Y.; Chudal, L.; Pandey, N.K.; Zhang, J.; Ma, L.; Xi, Q.; Yang, G.; Chen, Y.; et al. Use of copper-cysteamine nanoparticles to simultaneously enable radiotherapy, oxidative therapy and immunotherapy for melanoma treatment. Signal Transduct. Target. Ther. 2020, 5, 58. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Luo, J.; Xu, T.; Sun, K. N6-methyladenosine RNA modification in inflammation: Roles, mechanisms, and applications. Front. Cell Dev. Biol. 2021, 9, 670711. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhang, Y.; Li, D.; Cai, H.; Cai, L.; Xu, Q. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell. Signal. 2020, 69, 109553. [Google Scholar] [CrossRef] [PubMed]
- Hardbower, D.M.; Asim, M.; Luis, P.B.; Singh, K.; Barry, D.P.; Yang, C.; Steeves, M.A.; Cleveland, J.L.; Schneider, C.; Piazuelo, M.B.; et al. Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. USA 2017, 114, E751–E760. [Google Scholar] [CrossRef] [Green Version]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrocco, A.; Frawley, K.; Pearce, L.L.; Peterson, J.; O’Brien, J.P.; Mullett, S.J.; Wendell, S.G.; St Croix, C.M.; Mischler, S.E.; Ortiz, L.A. Metabolic Adaptation of Macrophages as Mechanism of Defense against Crystalline Silica. J. Immunol. 2021, 207, 1627–1640. [Google Scholar] [CrossRef]
- Rodriguez, A.E.; Ducker, G.S.; Billingham, L.K.; Martinez, C.A.; Mainolfi, N.; Suri, V.; Friedman, A.; Manfredi, M.G.; Weinberg, S.E.; Rabinowitz, J.D.; et al. Serine metabolism supports macrophage IL-1β production. Cell Metab. 2019, 29, 1003–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benada, O.; Pokorný, V. Modification of the Polaron sputter-coater unit for glow-discharge activation of carbon support films. J. Electron Microsc. Tech. 1990, 16, 235–239. [Google Scholar] [CrossRef]
- United Nations. The Globally Harmonized System of Classification and Labeling of Chemicals, Annex 10, Guidance Document on Transformation/Dissolution of Metals and Metal Compounds in Aqueous Media. Available online: https://unece.org/DAM/trans/danger/publi/ghs/ghs_rev07/English/13e_annex10.pdf (accessed on 24 March 2022).
- Hedberg, Y.; Midander, K.; Odnevall Wallinder, I. Particles, sweat, and tears: A comparative study on bioaccessibility of ferrochromium alloy and stainless steel particles, the pure metals and their metal oxides, in simulated skin and eye contact. Integr. Environ. Assess. Manag. 2010, 6, 456–468. [Google Scholar] [CrossRef]
- Mazinanian, N.; Hedberg, Y.; Odnevall Wallinder, I. Nickel release and surface characteristics of fine powders of nickel metal and nickel oxide in media of relevance for inhalation and dermal contact. Regul. Toxicol. Pharmacol. 2013, 65, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Hedberg, Y.S.; Odnevall Wallinder, I. Metal release from stainless steel in biological environments: A review. Biointerphases 2016, 11, 018901. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Herting, G.; Wei, Z.; Odnevall Wallinder, I.; Hedberg, Y. Bioaccessibility of nickel and cobalt in powders and massive forms of stainless steel, nickel-or cobalt-based alloys, and nickel and cobalt metals in artificial sweat. Regul. Toxicol. Pharmacol. 2019, 106, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Stirling, D.R.; Swain-Bowden, M.J.; Lucas, A.M.; Carpenter, A.E.; Cimini, B.A.; Goodman, A. CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinform. 2021, 22, 433. [Google Scholar] [CrossRef]
- Way, G. Blocklist Features—Cell Profiler. 2019. Available online: https://doi.org/10.6084/m9.figshare.10255811.v3 (accessed on 24 March 2022).
- Behdenna, A.; Haziza, J.; Azencott, C.A.; Nordor, A. pyComBat, a Python tool for batch effects correction in high-throughput molecular data using empirical Bayes methods. bioRxiv 2020. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]
- Chung, D.; Keleş, S. Sparse partial least squares classification for high dimensional data. Stat. Appl. Genet. Mol. Biol. 2010, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Chun, H.; Keleş, S. Spls: Sparse partial least squares (SPLS) regression and classification. In R Package, Version 2; 2012.
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alijagic, A.; Scherbak, N.; Kotlyar, O.; Karlsson, P.; Wang, X.; Odnevall, I.; Benada, O.; Amiryousefi, A.; Andersson, L.; Persson, A.; et al. A Novel Nanosafety Approach Using Cell Painting, Metabolomics, and Lipidomics Captures the Cellular and Molecular Phenotypes Induced by the Unintentionally Formed Metal-Based (Nano)Particles. Cells 2023, 12, 281. https://doi.org/10.3390/cells12020281
Alijagic A, Scherbak N, Kotlyar O, Karlsson P, Wang X, Odnevall I, Benada O, Amiryousefi A, Andersson L, Persson A, et al. A Novel Nanosafety Approach Using Cell Painting, Metabolomics, and Lipidomics Captures the Cellular and Molecular Phenotypes Induced by the Unintentionally Formed Metal-Based (Nano)Particles. Cells. 2023; 12(2):281. https://doi.org/10.3390/cells12020281
Chicago/Turabian StyleAlijagic, Andi, Nikolai Scherbak, Oleksandr Kotlyar, Patrik Karlsson, Xuying Wang, Inger Odnevall, Oldřich Benada, Ali Amiryousefi, Lena Andersson, Alexander Persson, and et al. 2023. "A Novel Nanosafety Approach Using Cell Painting, Metabolomics, and Lipidomics Captures the Cellular and Molecular Phenotypes Induced by the Unintentionally Formed Metal-Based (Nano)Particles" Cells 12, no. 2: 281. https://doi.org/10.3390/cells12020281