4.2. Classification of Andinia
The genus
Andinia has been recently revised and re-circumscribed by Wilson et al. [
15]. The authors described the genus as follows: “Plants caespitose or rhizomatous in habit; in the latter the rhizome repent, creeping or pendulous (similar to species of
Brachionidium). The inflorescence mostly successively multi-flowered, with only one flower open at a time. Ovaries glabrous to echinate. The flowers of some species similar to those of
Lepanthes. The petals mostly very much abbreviated compared to the sepals. The lip three-lobed (very shallowly in a few species), with the mid-lobe modified into an appendix in many species, and the lateral lobes frequently surrounding the column. Only a couple of species do not have an apical anther and stigma, but all have drop-like pollinaria, with a bubble-like viscidium”. We decided to cite expressis verbis Wilson’s et al. [
15] definition of
Andinia s.l. to make evident the problem in identification the unique combination of characters of this genus in its widest concept.
The habit detected in
Andinia s.l. is not unique at all, as was previously suggested by the authors. There can be distinguished two main habit types. The first one includes species with dangling rhizome, with short, pendulous ramicaul enclosed in typical, lepanthiform sheath(s) with ciliate or echinate margins. They produce orbicular to elliptic leaves, often with ciliate margins and/or upper surface. All of them have lepanthiform flowers with more or less connate sepals, and altered petals covered by various kinds of ciliae or hairs. The lip is simple in form, ecallose, canaliculated, and enclasping the gynostemium. The gynostemium is also lepanthiform, i.e., footless, terete, with erect anther lying on the dorsal surface of the column, and elongate, subulate rostellum. The stigmatic surface is subapical, horizontal, which facilitates grabbing the pollen mass. As thus can be seen, species of this group are similar to the representatives of
Lepanthes, and in fact for a long time all of them have been accommodated in this genus. Archila and Higgins [
13] were the first who emphasized differences between this group of species versus other
Lepanthes, giving them a rank of a separate genus (
Oreophilus W. Higgins & Archila), but for nomenclatural reason changing it soon to
Neooreophilus [
14]. Except for morphological background, the genus is supported by phylogenetic analyses in which all species belonging here create a monophyletic clade, although with various degrees of support.
Lepanthiform sheaths can be found also in
Lepanthes and
Trichosalpinx. Moreover, flowers of
Lepanthes and
Neooreophilus are similar in having basally connate sepals which are often widely spread and forming a kind of triangle, modified petals, and surface of various flower segments covered by ciliae or other protuberances. According to Wilson et al.’s [
15] phylogenetic analyses, however, these genera and
Neooreophilus are only distantly related. It is interesting to note that flowers of
Lepanthes–Trichosalpinx are borne below, whereas in
Neooreophilus they are above or upon the leaf blade. Phylogenetic studies can suggest independent origin of similar sheaths and other characters in both
Neooreophilus and
Lepanthes–Trichosalpinx. In our opinion it is necessary to consider as well a hybrid origin of the genus
Neooreophilus. The putative parents could be
Lepanthes and
Andinia species. It would be interesting to study sequences of other markers than
matK, ITS, or
rbcL in a wide spectrum of Pleurothallidinae.
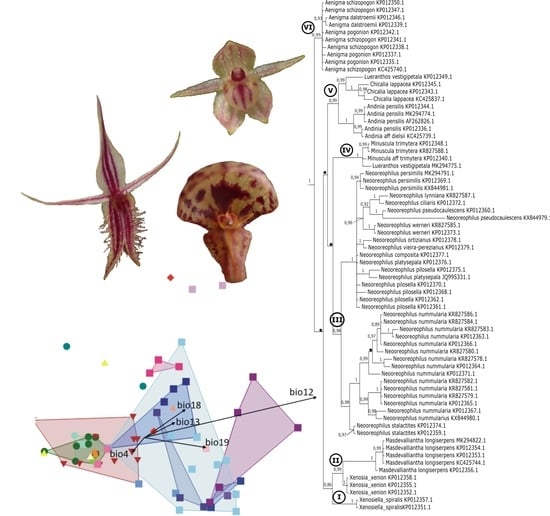
While
Neooreophilus was not well-sampled, several morphologically consistent groups can be recognized in the phylogenetic tree. The first subclade (B3) (
Figure 2,
Figure 3,
Figure 4,
Figure 5,
Figure 6 and
Figure 7) includes samples of species with ascending inflorescence borne above the leaf surface. Closely related
N. compositus,
N. persimilis, N. pilosellus, and
N. platysepalus are characterized by hirsute or ciliate–denticulate leaves and cucullate dorsal sepal. The group composed by
N. ciliaris and
N. lynnianius, which are characterized by glabrous leaves, is sister to
N. werneri, which has hirsute or ciliate–denticulate leaves and not cucullate dorsal sepals. The generitype of
Neooreophilus,
N. nummularius (B4) (
Figure 2,
Figure 3,
Figure 6 and
Figure 7), characterized by transversely ovate to trapeziform lip without an appendix, is sister to
N. stalactites (B5) (
Figure 2,
Figure 3,
Figure 6 and
Figure 7), which is the only representative of the genus having a bilaminate lip with an appendix.
The second large group of species is relatively uniform as the habit is considered and completely different from the aforementioned one. The plants are usually small to even tiny with short, creeping, or ascending rhizome. A short, erect ramicaul is enclosed in tight sheaths. Leaf is narrow, oblong to oblanceolate, always glabrous. Usually the few-flowered inflorescence is erect, rarely reclining, and longer than leaf. Both flower segments and reproductive structures are much more diversified than in Neooreophilus, and it is difficult to detect a common character to all of the species belonging here. This is not surprising, as according to the results of molecular analyses, they are paraphyletic, unlike Neooreophilus, which is monophyletic and strongly supported.
The clade B1 (
Figure 2,
Figure 3,
Figure 6 and
Figure 7) is much more complicated. It is represented in our study by
A. pensilis, A. dielsii, A. lappacea, and
A. vestigipetala. This clade is strongly supported on the trees obtained based on both ITS and combined matrices (
Figure 2,
Figure 3,
Figure 6 and
Figure 7), while on the cladograms performed for the
matK matrix, the relation between these taxa are unsolved (
Figure 4 and
Figure 5). In fact, it is very difficult to find a common character for such set of species. The only one we could detect are relatively small or even rudimentary petals. It is much easier to single out features characterized for each of them separately.
A. pensilis, to which we can also classified morphologically very similar
A. dielsii (a generitype of
Andinia) and the recently described
A. tingomariana, have a very peculiar gynostemium form, which can be compared to that found in
Neooreophilus and which could be described as lepanthiform, i.e., footless, erect, terete, with erect anther lying on its upper surface. The erect rostellum is much elongated, elliptic to oblong, and much exceeding the anther. The stigma is ventral. In all aforementioned species, the lip is basally (or even subapically in
A. tingomariana) connate with the gynostemium, more or less 3-lobed with both lateral lobes being upcurved and enclasping the gynostemium.
The subclade 1 (
Figure 6 and
Figure 7), including
A. pensilis and
A. dielsii, is sister to the pair of sample representatives
A. lappacea and
A. vestigipetala (
Figure 6 and
Figure 7), and it has both high value of probability (pp = 0.99) and bootstrap support (BS = 93). A similar situation we observed also on the trees preformed on the ITS marker (clade V,
Figure 2 and
Figure 3).
Andinia vestigipetala is unique in Pleurothallidinae in having erect, terete gynostemium, which is connate with rudimentary petals combined with simple, canaliculated lip, glabrous, free sepals, and glabrous ovary. A. lappacea is characterized by connate lateral sepals, and a subquadrate, ciliate lip, which is basally reflexed and apically emarginated. Sepals are densely ciliate–papillate. The gynostemium is short, clavate, slightly sigmoid, and basally enclasped by the lip. The ovary is echinate. We cannot find any mutual character for this pair except vestigial petals. The subclade A. pensilis, A. vestigipetala, and A. lappacea is weakly supported with Neooreophilus and A. trimytera.
However, based on the similarity in morphology of the flowers, the following species can be classified here as well: A. hystricosa, A. panica, A. sunchubambensis, and A. wayqechensis. All of them can be characterized by subsimilar sepals, of which both lateral ones are connate together in the lower half, prominent petals (except A. trimytera, where they are obscure), and peculiar lip. The lip is subsessile, strongly reflexed near the middle, with its basal part enclasping the gynostemium. Its apical part is deeply 3-lobed, with all lobes being subsimilar, more or less elongate. The central part of the lip is adorned by an elevated callus. The gynostemium is also uniform, i.e., footless, elongate, clavate, apically somewhat bent forward, and the apical clinandrium is obscure, the anther is apical and bent forward, and the rostellum is triangular–ligulate and bent over the stigmatic surface, which is ventral. The ovary is always hispid or muricate. All of these characters are shared by two additional species, A. pentamytera and A. ibex, despite some differences in their lip morphology (e.g., additional lobules in the sinus between middle and lateral lobes in the former, and two minute calli on the lip with widely spread lateral lobes in the latter one).
The taxa of group A (clade IV
Figure 2 and clade C
Figure 6) is represented by
A. pogonion,
A. schizopogon, and
A. dalstroemii. In our opinion, two other species should be classified with them, i.e.,
A. hirtzii and
A. uchucayensis, as they share many common characters. In all of them the ovary is echinate, lateral sepals are basally connate, petals are prominent, linear–lanceolate, acute to long–acuminate or even caudate, and the lip is sessile, gently reflexed near the middle, enclasping the gynostemium. The lip is suborbicular to elliptic–cordate, apically apiculate, or acute with central thickening. Its upper surface is more or less glandular or warty. The gynostemium is footless, rather short, clavate, somewhat sigmoid, the anther is suberect, the rostellum is short, suberect, and obtuse at the apex, and the ventral stigma is deeply concave. In all species of this group, inflorescence is few-flowered and usually longer than leaf, which is of course not a unique character, as it can be found elsewhere in
Andinia.
Andinia spiralis and
A. xenion have been classified to the genus
Xenosia by Luer (2004), and
A. longiserpens to
Masdevalliantha by Szlachetko and Margońska [
46].
Xenosia as originally described included three species, with
Pleurothallis xenion as the generitype.
Masdevalliantha included
M. masdevalliopsis and
M.
longiserpens only.
Xenosia as proposed by Luer [
47] appears to be a paraphyletic taxon in the light of molecular analyses. This impression is intensified by morphology. In the case of this clade we have been dealt a kind of puzzle. The genus
Xenosia can be characterized by a gynostemium structure, which is elongate, slender, clavate, slightly arcuate, with narrow apical clinandrium, apical, ventral anther, ligulate, incumbent rostellum, and short, baton-like column foot. This is the only mutual characteristic for the species of this genus, as they are different in other aspects of the flower structure. Inflorescence of
X. spiralis–X. macrorhiza is a few-flowered raceme, longer than leaf, with obtuse sepals, lateral sepals connate almost to the apex, sagittate lip apically papillate, with oblong, central callus. Inflorescence of
X. xenion is single-flowered, shorter than leaf, the sepals are caudate, free to the base, and the lip is basally 3-lobed with an oblong central callus. Its apical half is densely hairy. The gynostemium morphology and especially the connection between lip and column food recall the situation found in other
Xenosia species. The common character of
Masdevalliantha is a unique gynostemium morphology, which is short, massive, with long column foot and shelf-like, and massive process elevating rostellum. Especially interesting is the connection between lip and column foot, as the lip is attached on the ventral surface of the column foot, the apex of which is free and becoming accommodated by the basal lip cavity. We have not found such characteristics elsewhere in
Andinia s.l. species. Flower segments are similar to those found in
X. xenion, except lateral sepals, which in both
Masdevalliantha species are basally shortly connate.
Analysis of the distribution of all samples in the climatic space was carried out, where Andinia, Aenigma, Masdevalliantha, and Lueranthos were separated. The distribution of all observations in the two-dimensional climatic space indicates that the samples are widespread across the whole climatic space defined by the annual precipitation (bio12) and the temperature seasonality (bio4).
The question arises, what is the sense in creating genera which although monophyletic in terms of sequence of two molecular markers, are impossible to define in terms of morphology? The partial answer can be found in Chase et al. [
48]. The authors stated that their “general philosophy in developing the classification of Orchidaceae has been to minimize the number of tribes in order to make the system as simple as possible”. The next question is why they wish to minimize the number of tribes and other taxa? The answer is dumbfounding. They argued that “five is a reasonable number of subfamilies and is easily remembered by everyone”. This argument is difficult to challenge, but we dare to assume that if anyone is not able to remember six or seven subfamilies, that five is also too much to learn. Therefore Chase et al. [
48] “have tended to reduce well-supported sister tribes to a single tribe”. We can add that even such taxa are ill-defined. From the other papers they published, we can guess that it is a general rule they try to implement into the orchid classification on various taxonomic levels and is followed by other authors. In this case, it is less important whether the group can be defined in terms of morphology or not. Much more important is whether they are monophyletic in terms of one or another molecular marker, and it is completely unimportant whether it is possible to propose a key to determination of taxa distinguished in such an odd manner. Moreover, Górniak et al. [
39] was trying to prove that the topology of phylogenetic tree depends on the number of markers used in analyses and different taxonomic units could be proposed based on different tree topologies obtained.
To the best of our knowledge, the molecular marker used in analysis shows the phylogeny of this particular marker, and its extrapolation to the pattern of phylogeny of the entire organisms seems to be malfeasance. Furthermore, different markers can show different evolutionary paths which quite often are incongruent. In this case, which marker is more important?
Because all the above-mentioned morphologically uniform species groups form separated clades according to the results of phylogenetic analyses conducted by Wilson et al. [
15], we propose to split
Andinia s.l. into several genera, easily definable in terms of morphology and hence recognizable:
Aenigma,
Andinia,
Chicalia,
Lueranthos,
Masdevalliantha,
Minuscula,
Neooreophilus,
Xenosia, and
Xenosiella. Most of these taxa were previously considered as valid genera, and we believe that there is no reason to lump them into a single ill-defined genus. The taxonomic decisions presented here were made primarily on the basis of morphology, with DNA data demonstrating congruence with the morphological distinctions.
4.3. Taxonomic Treatment
Key to genera of Andinia-complex
1. Gynostemium footless or foot rudimentary … 2
1* Gynostemium with prominent column-foot … 6
2. Plants pendent, leaves appearing alternating and often overlapping … 1. Neooreophilus
2* Plants ascending or caespitose, leaves not appearing alternating and overlapping … 3
3. Inflorescence single-flowered … 4. Chicalia
3* Inflorescence few-flowered … 4
4. Petals prominent … 2. Aenigma
4* Petals much smaller than sepals … 5
5. Lip erect, canaliculate, petals rudimentary, minutely papillate along margins, fused with the base of the column part and tightly adnate to it … 5. Lueranthos
5* Lip and petals not as above … 3. Andinia
6. Ovary muricate … 6. Minuscula
6* Ovary glabrous ... 7
7. Inflorescence few-flowered … 9. Xenosiella
7* Inflorescence single-flowered … 8
8. Lip motile, base with prominent cavity accommodating free apex of the column foot … 7. Masdevalliantha
8* Lip deeply 3-lobed at the base, firmly fixed on the apex of the column foot … 8. Xenosia
1. Neooreophilus Archila
Revista Guatemalensis 12(2): 73. 2009; Generitype: Neooreophilus nummularius (Rchb.f.) Archila [≡ Lepanthes nummularia Rchb. f.]. ≡ Lepanthes sect. Brachycladae Rchb. f., Xenia Orchid. 1: 142. 1856. ≡ Lepanthes subgen. Brachycladium Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 15: 31. 1986. ≡ Oreophilus W. E. Higgins & Archila, Revista Guatemal. 12(2): 73. 2009, nom. illeg. ≡ Penducella Luer & Thoerle, Orchid Digest 74(2): 68. 2010, nom. illeg.
Plants pendent, delicate, with rarely branching rhizome, rooting occasionally. Ramicaul obscure, terete, enclosed in 1–2 lepanthiform sheaths. Leaf much longer than ramicaul, elliptic to suborbicular, hispid to ciliate all over, all glabrous, with prominent venation. Few-flowered inflorescence usually shorter than leaf. Flowers typically lepanthiform, with sepals dissimilar or subsimilar in form and size, variously connate, lateral ones sometimes almost completely united together, more or less papillate or ciliate. Petals distinctly smaller than sepals, sometimes rudimentary, variously formed, longer than wide, or wider than long, entire or lobed, ciliate, papillate or with other protuberances. Lip shortly clawed to sessile, ecallose, with more or less auriculate base, lateral lobes upcurved, enveloping the gynostemium, ciliate or papillate. Gynostemium short, erect, terete, column foot missing, stigma subapical, horizontal, rostellum elongate, subulate or digitate, much exceeding the anther, erect or upcurved, anther erect, dorsal, ellipsoid, pollinia clavate, viscidium large, plate-like.
Three clearly separated assemblages in Neooreophilus can be distinguished based on morphological data and outcomes of molecular analyses, and these groups are presented here as subgenera. To facilitate identification of representatives within the largest subgenus, Amplectentes, this taxon was further divided into sections. However, it is important to emphasize the poor sampling of Neooreophilus that complicates comparison of morphological data with phylogenetic relationships. Based on similarity of related species, the more complicated division of the genus could be conducted based, for example, on petal form. More comprehensive molecular study is required to propose formal sectional division of the largest subgenus of Neooreophilus.
Key to subgenera of Neooreophilus
1. Inflorescence reclining, borne upon the leaf surface … 2
1* Inflorescence ascending, borne above the leaf surface … 1.1. Neooreophilus subgen. Amplectentes
2. Lip bilaminate with an appendix …. 1.2. Neooreophilus subgen. Bilamellatae
2* Lip transversely ovate to trapeziform in outline, without an appendix ... 1.3. Neooreophilus subgen. Neooreophilus
1.1. Subgenus Amplectentes (Luer) Kolan. & Szlach., stat. et comb. nov.
Basionym: Lepanthes sect. Amplectentes Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 52: 3. 1994. ≡ Andinia sect. Amplectentes (Luer) Karremans & S. Vieira-Uribe, Phytotaxa 295(2): 123. 2017. Type: Lepanthes pilosella Rchb. f., Flora 69: 556. 1886.
This is the largest subgenus of Neooreophilus. Three species groups can be distinguished within this subgenus.
Neooreophilus ciliaris-group
This group is characterized by glabrous leaves. Species included in this group:
Neooreophilus ariasianus (Luer & L.Jost) Archila, Revista Guatemalensis 12(2): 73 (-74). 2009.
Neooreophilus bifidus Tobar & Archila, Revista Guatemalensis 15(2): 2,
Figure 1 and
Figure 2. 2012.
Neooreophilus chaoae S.V.Uribe & L.Jost, Lankesteriana 15(3): 213. 2015.
Neooreophilus chelosepalus (Luer & Hirtz) Archila, Revista Guatemalensis 12(2): 76. 2009.
Neooreophilus ciliaris (Luer & Hirtz) Archila, Revista Guatemalensis 12(2): 76. 2009.
Neooreophilus cordilabius (Luer) Archila, Revista Guatemalensis 12(2): 77. 2009.
Neooreophilus dentatus Archila, Revista Guatemal., 17(1): 41. 2014.
Neooreophilus destitutus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 78. 2009.
Neooreophilus lunaris (Luer) Archila, Revista Guatemalensis 12(2): 81. 2009.
Neooreophilus lynnianus (Luer) Archila, Revista Guatemalensis 12(2): 81. 2009.
Neooreophilus macroticus (Luer & Dalström) Archila, Revista Guatemalensis 12(2): 82. 2009.
Neooreophilus ortizianus S.V.Uribe & Thoerle, Orquideologia 28(2): 135 (-139; fig.; photogr.). 2011.
Neooreophilus pendens (Garay) Archila, Revista Guatemalensis 12(2): 84. 2009.
Neooreophilus pholeter (Luer) Archila, Revista Guatemalensis 12(2): 85.
Neooreophilus ricii (Luer & R.Vásquez) Archila, Revista Guatemalensis 12(2): 86. 2009.
Neooreophilus triangularis (Luer) Archila, Revista Guatemalensis 12(2): 87. 2009.
Neooreophilus vieira-perezianus P.Ortiz, Orquideologia 28(1): 7 (-10;
Figure 1; photogr.). 2011.
1.1.2. Neooreophilus pilosellus group
This group is characterized by hirsute or ciliate–denticulate leaves and cucullate dorsal sepal. It includes:
Neooreophilus cardiochilus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 75. 2009.
Neooreophilus chilopsis (Luer & Hirtz) Archila, Revista Guatemalensis 12(2): 76. 2009.
Neooreophilus compositus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 77. 2009.
Neooreophilus erepsis (Luer & Hirtz) Archila, Revista Guatemalensis 12(2): 78. 2009.
Neooreophilus lunatocheillus Tobar & Archila, Revista Guatemalensis 15(2): 26,
Figure 3. 2012.
Neooreophilus mongei Tobar & Archila, Revista Guatemalensis 15(2): 23,
Figure 2. 2012.
Neooreophilus octocornutus (Luer) Archila, Revista Guatemalensis 12(2): 84. 2009.
Neooreophilus persimilis (Luer & Sijm) Archila, Revista Guatemalensis 12(2): 84. 2009.
Neooreophilus phalicus Tobar & Archila, Revista Guatemalensis 15(2): 20,
Figure 1. 2012.
Neooreophilus pilosellus (Rchb.f.) Archila, Revista Guatemalensis 12(2): 85. 2009.
Neooreophilus platysepalus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 85. 2009.
Neooreophilus tridactylus (Luer) Archila, Revista Guatemalensis 12(2): 88. 2009.
Neooreophilus ursulus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 88. 2009.
Neooreophilus viebrockianus (Luer & L.Jost) Archila, Revista Guatemalensis 12(2): 89. 2009.
Neooreophilus villosus (Løjtnant) Archila, Revista Guatemalensis 12(2): 89. 2009.
Neooreophilus medinae Kolan. & Szlach., sp. nov. TYPE: Colombia. Dept. Putumayo. Vereda Balsayaco, near San Andres. Alt. 1984 m. 7 September 2016. R. Medina T. S14/28 (holotype, JAUM!; MEDEL!—photos).
[urn:lsid:ipni.org:names:xxxxxxx-x]
Species similar to N. platysepalus, N. ursulus, and N. pilosellus, distinguished by relatively shortly connate lateral sepals and prominent lip auricles. Moreover, from N. platysepalus and N. pilosellus, the new entity differs additionally by the lip and externally densely ciliate dorsal sepal.
Epiphytic plant, up to 15 cm long in total; ramicauls spaced 4–5 mm, each segment enclosed by two imbricating, membranaceous, infundibular sheaths, ciliate along the ribs, with dilated, ciliated ostia. Ramicauls 0.8–1.1 mm long, enclosed by a single membranaceous, infundibular sheath ciliate along the ribs, with dilated, ciliate ostia. Leaves shortly petiolate; petiole 0.7–1 mm long; blade 9–10 mm long, 3.5–4 mm wide, elliptic to ovate-elliptic, subacute to acute, densely ciliate, 3-nerved, lateral veins sometimes dichotomous. Inflorescence borne upon the surface of leaf, single-flowered; floral bract about 0.5 mm long, infundibular, oblique, acuminate, membranaceous; pedicel slender, persistent, 1.5–2 mm long; ovary ciliate, about 1 mm long. Flower small, sepals light yellow with dark red stripes, dorsal sepal mottled with purple along veins, petals light yellow with purple stripe, lip light yellow with 3 purple veins. Dorsal sepal concave in the natural position, 7.5 × 6 mm when expanded, broadly obovate, with a small, obtuse apiculus, 5- or 7-veined, externally densely ciliate. Lateral sepals 7 × 4 mm, connate for about 4 mm, narrowly ovate, obtuse, 2-veined, externally glabrous. Petals 2–2.1 × 0.4 mm, oblong–lanceolate to linear–lanceolate, basally cuneate, obtuse, 1-veined, glabrous. Lip embracing the column in the natural position, 1.7 × 1.5 mm when expanded, subsessile, lamina subrectangular with prominent basal projections 0.7 × 0.5 mm, apical part truncate with a small, obtuse apiculus and here glandular–ciliate, margin glabrous, disc 3-veined, glabrous. Gynostemium about 2 mm long, terete, anther and stigma apical (
Figure 13 and
Figure 14).
Etymology: Dedicated to Ramiro Medina Trejo, an orchid enthusiast who collected and cultivated new species.
Distribution, habitat and ecology: So far, Neooreophilus medinae is known only from the type locality in southern Colombia, where it was found on the mossy trunk and large branches of the tree in the pasture at the altitude of almost 2000 m. The population consists of about 50 specimens and its individuals grow sympatrically with specimens of N. pseudocaulescens. Flowering occurs in September.
Notes. Neooreophilus medinae resembles Colombian N. platysepalus, N. pilosellus, and N. ursulus, but the prominent basal projections of the lip allow one to easily distinguish it from the aforementioned species. In both N. pilosellus and N. ursulus, the lateral sepals are long-connate, free only at the apices. The most similar species is N. platysepalus known from the Colombian department of Antioquia, which is characterized by the glabrous sepals and small lip that is wider than long (1–1.25 × 1.5 mm), subquadrate to transversely subcordate, rounded to subtruncate at the apex, and minute basal projections. The lateral sepals of this species are connate in the basal 3/4–4/5.
Conservation status. Considering that about 50 specimens were found, the threat category according to IUCN [
49] criterion D swings between Critically Endangered (CR) and Endangered (EN), while for the definition of the category according to criterion B data are missing on possible decline and fluctuations. Therefore, more in field research would be useful for the definition of distribution, population size, locations (
sensu IUCN) and threats. In fact, new described species with a restricted distribution could be attributed to different IUCN categories, as Critically Endangered (CR) according to criterion B [
50], or Vulnerable (VU) under criterion D [
51,
52].
1.1.3. Neooreophilus werneri-group
This group is characterized by hirsute or ciliate–denticulate leaves, not cucullate dorsal sepal, and variously connate, spreading lateral sepals. It includes:
Neooreophilus catellus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 75. 2009.
Neooreophilus dactylus (Garay) Archila, Revista Guatemalensis 12(2): 78. 2009.
Neooreophilus exiguus (Luer & Hirtz) Archila, Revista Guatemalensis 12(2): 79. 2009.
Neooreophilus geminipetalus (Luer & J.Portilla) Archila, Revista Guatemalensis 12(2): 79. 2009.
Neooreophilus hippocrepicus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 80. 2009.
Neooreophilus irrasus (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 80. 2009.
Neooreophilus lueri (S.V.Uribe & Karremans) Kolan. & Szlach., comb. nov.
Basionym: Andinia lueri S.V.Uribe & Karremans, Orquideologia 33: 116. 2016.
Neooreophilus lupulus (Luer & Hirtz) Archila, Revista Guatemalensis 12(2): 81. 2009.
Neooreophilus micropetalus (L.O.Williams) Archila, Revista Guatemalensis 12(2): 82. 2009.
Neooreophilus monilius (Luer & R.Escobar) Archila, Revista Guatemalensis 12(2): 83. 2009.
Neooreophilus montis-rotundus (P.Ortiz) Archila, Revista Guatemalensis 12(2): 83. 2009.
Neooreophilus pseudocaulescens (L.B.Sm. & S.K.Harris) Archila, Revista Guatemalensis 12(2): 86. 2009.
Neooreophilus rotundus Archila, Revista Guatemal. 17(1): 42. 2014.
Neooreophilus sibundoyensis Kolan., Ann. Bot. Fenn. 50(3): 170. 2013.
Neooreophilus sudamericanus Archila, Revista Guatemal., 17(1): 40. 2014.
Neooreophilus werneri (Luer) Archila, Revista Guatemalensis 12(2): 89. 2009.
Incertæ sedis
Due to the lack of sufficient information about morphological characters of the following species we were not able to assign them into any of the groups listed above:
Neooreophilus auriculatus Archila, Revista Guatemal. 17(1): 44. 2014.
Neooreophilus caveroi (D.E. Benn. & Christenson) Archila, Revista Guatemal. 12(2): 76. 2009.
Neooreophilus roseus Archila, Revista Guatemal. 17(1): 43. 2014.
1.2. Subgenus Bilamellatae (Luer) Kolan. & Szlach., stat. et comb. nov.
Basionym: Lepanthes sect. Bilamellatae Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 52: 3. 1994; Type: Lepanthes stalactites Luer & Hirtz, Lindleyana 2(2): 105. 1987.
This group is characterized by the inflorescence reclining, borne upon the leaf surface and bilaminate lip with an appendix.
1.3. Subgenus Neooreophilus
This group is characterized by the inflorescence reclining, borne upon the leaf surface and transversely ovate to trapeziform lip without an appendix.
Neooreophilus nummularius (Rchb.f.) Archila, Revista Guatemalensis 12(2): 83. 2009.
Neooreophilus obesus (S.V.Uribe & Karremans) Kolan. & Szlach., comb. nov.
Basionym: Andinia obesa S.V.Uribe & Karremans, Lankesteriana 17: 311.2017.
2. Aenigma (Luer) Szlach. & Kolan., stat. et comb. nov.
Basionym: Pleurothallis subgen. Aenigma Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 20: 26. 1986. ≡ Andinia subgen. Aenigma (Luer) Karremans & M. Wilson, Phytotaxa 295(2): 121. 2017. Generitype: Aenigma schizopogon (Luer). Szlach. & Kolan. [≡ Pleurothallis schizopogon Luer].
All species small, with somewhat elongate and ascending rhizome, producing ramicauls in short distance. Ramicaul short, terete, enveloped in 1 tight sheath. Leaf oblong, glabrous. Inflorescence longer than leaf, few-flowered, with flowers opening in succession. Ovary muricate, pedicel glabrous. Lateral sepals connate in the lower half, more or less acuminate, glabrous, or hispid. Petals prominent, linear–lanceolate, acuminate, as long as sepals, but distinctly narrower. Lip sessile, firmly connate with the column foot, more or less reflexed just above the base, enveloping the gynostemium, with prominent calli in the center, suborbicular to pentagonal in outline, shortly apiculate in the apex. Gynostemium erect to somewhat arcuate, clavate, column foot rudimentary, stigma subapical, rostellum blunt, short, rounded at the apex, anther subapical to subventral, suberect, ovoid–ellipsoid. The genus includes:
Basionym: Pleurothallis dalstroemii Luer, Orchideer 5: 52. 1984.
Basionym: Andinia hirtzii Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 103: 275. 2005.
Basionym: Pleurothallis pogonion Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 52: 61. 1994.
Basionym: Pleurothallis schizopogon Luer, Lindleyana 16(4): 251. 2001.
Basionym: Andinia uchucayensis A.Doucette & J.Portilla, Orchids (Lindleyana) 86(1): 72. 2017.
3. Andinia (Luer) Luer
Monogr. Syst. Bot. Missouri Bot. Gard. 79: 5. 2000. ≡ Salpistele subgen. Andinia Luer, (Monogr. Syst. Bot. Missouri Bot. Gard. 39: 124. 1991; Generitype: Andinia dielsii (Mansf.) Luer [≡ Lepanthes dielsii Mansf.].
Plants with elongate rhizome, producing ramicauls in prominent intervals. Ramicaul short, terete, enclosed in 1–2 tight sheaths. Leaf oblong elliptic, glabrous. Multiflowered inflorescence much longer than leaf. The flowers produced in succession, broadly opened. Ovary muricate, or glabrous, pedicellate. Sepals subsimilar in size and form, shortly acuminate, mucronate, ciliate, or glabrous along margins, lateral sepals connate in the lower half or free to the base. Petals much smaller than sepals. Lip simple, 3-lobed, basally or subapically united with the gynostemium, lateral lobes upcurved, enveloping the gynostemium, with prominent callus in the center or ecallose. Gynostemium erect, slender, terete, column foot missing, stigma ventral, emarginate, anther subdorsal, erect, ovoid, rostellum elongate, ligulate, erect, much exceeding the anther.
The genus includes:
Andinia dielsii (Mansf.) Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 79: 6. 2000.
Andinia pensilis (Schltr.) Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 79: 6. 2000.
Andinia tingomariana A.G. Diaz & Mark Wilson, Phytotaxa 361(2): 223–227. 2018.
4. Chicalia Szlach. & Kolan., gen. nov.
Generitype: Chicalia lappacea (Luer) Szlach. & Kolan. [≡ Pleurothallis lappacea Luer]
Etymology: In reference to Chical, Carchi, Ecuador, where the type specimen of P. lap-pacea was collected.
Plants very small, subcaespitose. Ramicaul obscure, enclosed in 1 tight sheath. Leaf elliptic, glabrous. Single-flowered inflorescence longer than leaf, produces relatively large, broadly opened flower. Ovary densely muricate, pedicellate. Sepals subsimilar in size, papillate all over except central and basal parts, lateral sepals connate together almost to the apex forming a kind of platform, supporting lip. Petals rudimentary, with ciliate margins. Lip sessile, subquadrate in outline, reflexed just above the base, enveloping partially the gynostemium, with prominently emarginate apex. Its surface ciliatae–glandular, except the central and apical portions. Gynostemium rather short, clavate, subsigmoid, with obscure column foot, anther apical, incubment ellipsoid, rostellum subapical, short, obscure, stigma subapical.
The genus includes:
Basionym: Pleurothallis lappacea Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 79: 129, f. 5. 2000.
5. Lueranthos Szlach. & Marg.
Polish Bot. J. 46(2): 117. 2001 [2002]; Generitype: Lueranthos vestigipetalus (Luer) Szlach. & Marg. [≡ Pleurothallis vestigipetala Luer].
Plants with elongate, ascending rhizome. Ramicaul short, terete, enclosed in tight sheath. Inflorescence few-flowered, somewhat longer than leaf. Flowers produced in succession with glabrous ovary and broadly opened sepals free to the base. Sepals similar in size and form. Petals rudimentary, minutely papillate along margins, fused with the base of the column part and tightly adnate to it. Lip erect, canaliculate, oblong–elliptic, ecallose, simple, more or less papillate all over. Gynostemium erect, terete, slender, elongate, footless, with subapical, incumbent anther and short rostellum. Anther oblong ovoid. Stigma apical, horizontal.
This monotypic taxon includes:
6. Minuscula Szlach. & Kolan., gen. nov.
Generitype: Minuscula trimytera (Luer & R.Escobar) Szlach. & Kolan. [≡ Pleurothallis trimytera Luer & R. Escobar].
Etymology: From minusculus (Lat.)—small, tiny. An allusion to the size of species of this group.
Plants miniature, with very short, creeping rhizome or caespitose. Ramicaul obscure, terete, enclosed in 1 tight sheath. Leaf elliptic, shortly apiculate, glabrous. The inflorescence longer than leaf; 1-(2)-flowered. Flower broadly opened. Ovary muricate, pedicellate. Sepals subsimilar in form and size, glabrous, papillate or glandular in the basal part or all over, lateral sepals connate in the lower half, occasionally free. Petals shorter and narrower than sepals, occasionally rudimentary. Lip shortly clawed, transversely elliptic, deeply 3-lobed at the apex, geniculately bent forward, thickened in the center, concave below, rarely calli obscure. Gynostemium slightly sigmoid, clavate, column foot short, apical clinandrium obscure, anther apical, incumbent, oblong ovoid, stigma ventral, rostellum ventral, triangular, more or less incumbent.
The genus includes:
Basionym: Pleurothallis hystricosa Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 52: 54. 1994.
Basionym: Pleurothallis ibex Luer, Selbyana 5(2): 168 (–169). 1979.
Basionym: Pleurothallis panica Luer & Dalström, Monogr. Syst. Bot. Missouri Bot. Gard. 61(3): 6. 1996.
Basionym: Pleurothallis pentamytera Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 52: 58. 1994.
Basionym: Andinia sunchubambensis A. Doucette & Janovec, Internet Orchid Sp. Photo Encycl. Nomencl. Notes 2016: f. 1A–G, 2–3. 2016.
Basionym: Pleurothallis trimytera Luer & R. Escobar, Orquideología 16: 34. 1983.
7. Masdevalliantha (Luer) Szlach. & Marg.
Polish Bot. J. 46(2): 117. 2001. ≡ Pleurothallis subgen. Masdevalliantha Luer, Monogr. Syst. Bot. Missouri Bot. Gard. 20: 44. 1986. ≡ Andinia subgen. Masdevalliantha (Luer) Karremans & M. Wilson, Phytotaxa 295(2): 125. 2017; Generitype: Masdevalliantha masdevalliopsis (Luer) Szlach. & Marg. [≡ Pleurothallis masdevalliopsis Luer].
Subcaespitose plant, with 1-flowered inflorescence longer than leaf. Ramicaul short, terete, with tight sheath. Flowers broadly opened. Sepals subsimilar in form and size, long–caudate, lateral sepals connate basally. Petals orbicular to obtriangular, much smaller than sepals. Lip motile, more or less 3-lobed, base with prominent cavity accommodating free apex of the column foot, apical part of the middle lobe hairy or hispid, callus single, oblong, or bifid. Gynostemium short, massive, suberect, rostellum elongate, oblong–triangular, perpendicular to the column axis, column foot long, upcurved with the lip attached below its apex, which is placed in the cavity at the lip base. Anther subapical, suberect, ovoid, relatively small. Stigma ventral.
The genus includes:
Masdevalliantha longiserpens (C. Schweinf.) Szlach. & Marg., Polish Bot. J. 46(2): 117. 2001.
Masdevalliantha masdevalliopsis (Luer) Szlach. & Marg., Polish Bot. J. 46(2): 117. 2001.
8. Xenosia Luer
Monogr. Syst. Bot. Missouri Bot. Gard. 95: 265. 2004; Generitype: Xenosia xenion (Luer & R. Escobar) Luer [≡ Pleurothallis xenion Luer & R. Escobar].
Etymology: In reference to the similarity to the genus Xenosia.
Plants miniature. Rhizome shortly repent. Ramicaul filiform, tightly enclosed in 1 sheath. Leaf linear–lanceolate, glabrous. Inflorescence shorter than leaf, single-flowered. Flowers broadly opened. Ovary glabrous, pedicellate. Sepals subsimilar in form and size, free to the base, long–caudate. Petals deltoid, prominently smaller than sepals. Lip deeply 3-lobed at the base, firmly fixed on the apex of the column foot, with basal lateral lobes obliquely rhombic, and oblong callus in the center, the middle lobe densely pubescent. Gynostemium slender, elongate, terete, apically somewhat arcuate. Apical clinandrium narrow. Column foot elongate, robust. Anther apical, incumbent, ovoid. Rostellum ventral, ligulate, incumbent. Stigma ventral.
The genus includes one species:
9. Xenosiella Szlach. & Kolan., gen nov.
Generitype: Xenosiella spiralis (Ruiz & Pav.) Szlach. & Kolan. [≡ Humboltia spiralis Ruiz & Pav.]
Plants with elongate, ascending rhizome. Pseudobulbs terete, short, enclosed in tight sheath. Leaf ligulate, glabrous. Inflorescence shorter than leaf, few-flowered. Flowers tubular borne in succession. Ovary glabrous, pedicellate. Sepals subsimilar in size and form, obtuse, glabrous, lateral sepals connate together in the basal 2/3, forming a kind of platform below lip. Petals oblong–obovate, much smaller than sepals. Lip firmly attached to the column foot, deltoid–ovate in outline, canaliculate, obscurely 3-lobed, with oblong callus in the center and papillate apical lobe. Gynostemium elongate, slender, clavate, with prominent, slightly upcurved column foot. Anther ventral, ellipsoid, pollinia 2, ovoid. Rostellum ligulate, obtuse, incumbent. Viscidium plate-like. Stigma ventral.
The two species classified in this genus are probably synonymic as the only noticeable difference between them is the lip base (cuneate in X. spiralis and rounded-broadly cuneate in X. macrorhiza). The genus includes:
Basionym: Pleurothallis macrorhiza Lindl., Monogr. Syst. Bot. Missouri Bot. Gard. 105: 233–234, f. 185. 2006.
Basionym: Humboltia spiralis Ruiz & Pav., Syst. Veg. Fl. Peruv. Chil. 1: 237. 1798.