Editorial: Cellular Mechanisms During Normal and Abnormal Craniofacial Development
- 1Department of Biological Sciences, SUNY Oswego, Oswego, NY, United States
- 2Department of Natural Sciences, University of Michigan–Dearborn, Dearborn, MI, United States
- 3Department of Experimental Biology, Faculty of Science, Masaryk University, Brno, Czechia
- 4Laboratory of Molecular Morphogenesis, Institute of Animal Physiology and Genetics, Czech Academy of Sciences, Brno, Czechia
Editorial on the Research Topic
Cellular Mechanisms During Normal and Abnormal Craniofacial Development
Introduction
Embryonic craniofacial development involves a series of cellular processes that drive patterning, outgrowth, and fusion of a number of independently forming components. The progression of growth and morphogenesis relies on cellular mechanisms such as differentiation, proliferation, migrations, transformation, and apoptosis to form the correct shape and structure in the developing embryo (Figure 1). During craniofacial development, the aforementioned processes are spatiotemporally constrained, allowing for multiple mechanisms within a relatively small region in order to create complex and intricate structures. Failure at any stage risks considerable consequences for the embryo, ranging from slight defects in craniofacial patterning to total inviability.
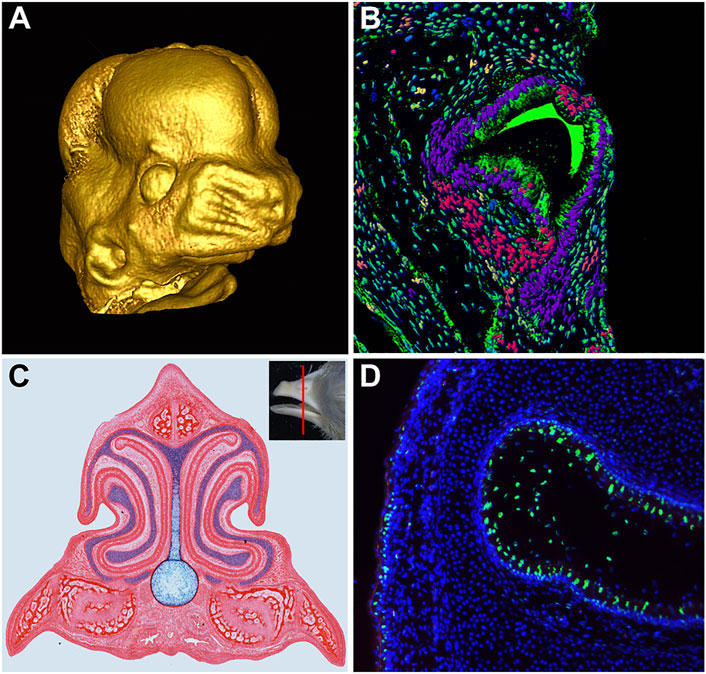
FIGURE 1. Development of Craniofacial Structures. (A) MicroCT scan of embryonic day 13 mouse embryo face, reconstructed using MicroView software. (B) Transverse section of the chameleon tooth. Artificial colors of nuclei were produced in software IMARIS followed by segmentation and analysis for cell nuclei shape differences. (C) Transverse section of Hamburger and Hamilton (1951) stage 40 chicken embryo beak stained with picrosirius red to highlight skeletal and soft tissue and alcian blue to highlight cartilage. (D) Apoptotic signal (green) in the developing nasal cavity of a Hamburger and Hamilton (1951) stage 42 chicken embryo.
The current Research Topic “Cellular Mechanisms During Normal and Abnormal Craniofacial Development” intends to examine and collate the most up-to-date studies on the cellular processes that drive embryonic craniofacial morphogenesis. In recent years, scientists have taken great strides towards attaining a better understanding of the cellular mechanisms involved in the fundamental aspects of craniofacial development. Seemingly disparate fields such as computer sciences, mathematics, tomographic and microscopic imaging, biochemistry, and molecular biology have come together to greatly enhance our understanding of how individual cells influence the overall patterning and morphogenesis of the embryonic face.
This Research Topic includes 21 papers that focus on the cellular processes involved in the development of a variety of craniofacial structures such as palate, teeth, eyes, craniofacial muscles, as well as their contribution to associated developmental defects. Additionally, the importance of interaction between neural tissue and forming face is introduced and discussed.
Cellular Processes in Craniofacial Structures Shaping and Patterning
Murrilo-Rincón and Kaucka reviewed the complexity of craniofacial structure development from a cellular perspective. They focused on the cell fate specification in craniofacial structures from the emergence of neural crest cells up to the establishment of facial geometry. Individual or collective cell migration, cellular interactions, and physical forces generated by morphogenetic events, all contribute to the establishment of the face as well as intra- and inter-species morphological variability. The importance of molecular regulation, including epigenetic events and alteration of signaling center activity, is introduced, in addition to the different types of craniofacial defects they can lead to, ranging from subtle changes of facial symmetry to severe lethal conditions.
In vertebrates, morphogenesis of the upper jaw and face is regulated by cellular interactions between the brain, facial ectoderm, and neural crest mesenchyme; mediated by several morphogenetic signals including wingless (WNT) (Geetha-Loganathan et al., 2009; Rickbourg et al., 2020), sonic hedgehog (SHH) (Hu et al., 2015), fibroblast growth factor (FGF) (Hu et al., 2003), and bone morphogenetic protein (BMP) (Barlow and Francis-West, 1997). During embryonic development, the relationship between brain and face has also received some attention, with disruption of signals altering patterning of both structures (Richbourg et al., 2020).
Since Wnt signaling from the facial ectoderm and forebrain is required to regulate the outgrowth of facial prominences, activating and inhibiting the expression of canonical Wnt signaling in these regions causes malformation of facial structures, including cleft lip (Green et al., 2019). In this topic, Marchini et al. illustrate the interactions between the brain and face by activating WNT3a and inhibiting Wnt signaling with DKK1 in the neural crest of developing chicken embryos. Their study uncovered that WNT3a activation expands the shape of the frontonasal ectodermal zone (FEZ) along with the brain and face during early face formation, whereas changes in the brain and face are decoupled during later developmental stages, supporting the palimpsest model for morphological integration (Hallgrímsson et al., 2009).
Mutations in Ras-related C3 botulinum toxin substrate 1 (Rac1) encoding a small GTPase have been shown to be involved in brain tumorigenesis and abnormal development (Khalil and El-Sibai, 2012). Gahankari et al. have elucidated the activity of Rac1 signaling during the development of the midbrain dopaminergic neurons (mDA). They have created Rac1CA;Wnt1-Cre2 mutant mice with specific activation of Rac1G12v mutant in neural crest cell derivatives. These mutants are characterized by an enlarged midbrain due to increased proliferation and abnormal migration of neural cells, disrupting localization of mDA in the ventral side.
Palatogenesis
In the mammalian embryo, the assembly of the maxillary region is a precarious period of development. Improper fusion of the upper lip and/or secondary palate is exceedingly common, with the incidence of cleft lip/palate (CLP) in humans occurring at a rate of one in 750 newborns, among the most frequent birth defects worldwide (Kousa and Schutte, 2016). Palate development consists of the growth, elevation, and fusion of the palatal shelves, starting at 6–8 weeks of gestation in humans (Mossey et al., 2009). Cleft palate (CP) is etiologically complex and heterogeneous, occurring in isolated and syndromic cases, and involving a combination of genetic and environmental factors, with many cases remaining unexplained (Mossey et al., 2009; Buser and Pohl, 2015).
Manuscripts in the recent topic focused on a wide range of developmental processes ranging from undersized palatal shelves to the retention of the medial epithelial seam (MES). One of the disorders associated with cleft palate is KGB syndrome. The suspected cause of KGB syndrome is the Ankrd11 gene, a chromatin regulator associated with neural stem cell fates. Here, Roth et al. report that mice with heterozygous deletion of Ankrd11 in neural crest cells display craniofacial defects that mirror KBG syndrome, including hypoplastic palatal shelves, which the study found was due to a regional decrease in mesenchymal proliferation. Ultimately, their study identifies Ankrd11 as a critical regulator of intramembranous ossification and palate development and suggests that Ankrd11 neural crest knockout mice may serve as novel models for KBG syndrome.
Keeping the focus on cell proliferation in palatal shelves, Yoshioka et al. studied the effect of all-trans retinoic acid (atRA) on microRNA (miR) expression during palatogenesis. atRA is used in the treatment of skin diseases and cancers, and yet is a teratogen known to cause cleft palate. The authors found that excessive atRA induces expression of MicroRNA-4680-3p, which suppresses genes crucial for palate development, leading to decreased cell proliferation in human embryonic palatal mesenchymal cells. This study is important in not only highlighting the involvement of miRs in palatogenesis but also elucidating a molecular relationship between miRs and palatal clefting.
In case shelves reach an appropriate size, they have to undergo elevation from a vertical position to horizontal; yet another potential pitfall. On occasion, palatal shelves fail to horizontalize in a timely manner. Bukova et al. observed this phenotype in their study of the CRISPR/Cas9 WIZ knockout mouse model. The authors found that loss of Wiz causes disruption of histone methylation known to be driven by the G9a/GLP methylation complex. Altered histone methylation causes delayed palatal shelf horizontalization, which resulted in CP since the medial edge epithelium of the palatal shelves misses the window of competence to fuse, even if horizontalization eventually occurs.
Even after the horizontalization and approach of palatal shelves, the fusion process is complex and often disrupted. Van der Woude syndrome (VWS) is associated with palatal clefting; with almost three-quarters of cases caused by mutations in the transcription factor IRF6. IRF6 is a master regulator balancing keratinocyte proliferation and differentiation, with Irf6-deficient mice displaying disorganized oral epithelium. In their study, Degen et al. identified a novel heterozygous VWS-causing IRF6 mutation that lacks part of its protein-binding domain and entire C-terminus. This truncated IRF6 copy undergoes nonsense-mediated mRNA decay, leading to haploinsufficiency. The remaining IRF6 copy can still control gene regulatory networks but does not meet the threshold required for proper periderm arrangement, resulting in pathological epithelial fusions between tissues in the oral cavity, thereby interfering with shelf lift, outgrowth, and fusion.
In studying the mechanism of IRF6-caused CP, Girousi et al. found that lack of IRF6 disrupts epithelial homeostasis by altering colony morphology, migration pattern, and differentiation potential of cultured postnatal skin- and oral mucosa-derived keratinocytes. Colonies appeared scattered with less stable cell-cell contacts and a significant number of single and enlarged cells. Wound-healing experiments showed IRF6-deficient keratinocytes preferentially moving as single cells, with random and undirected migration, potentially explaining the appearance of disrupted epithelium in IRF6 mutants. The proteomic analysis further supported these results by finding that most of the differentially expressed proteins in the absence of IRF6 were associated with differentiation, cell-cell adhesion, and immune response.
Assuming epithelia do manage to fuse, the disintegration of the MES needs to occur in order for fusion to be complete. Verheijen et al. studied the interplay between CXCL12 ligand and CXCR4 receptor in association with the breakdown of the MES and osteogenesis during palatal fusion in mice. They conclude that CXCL12 and Sox9 expression by the disintegrating MES and osteogenic centers in the fusing palatal shelves could promote the maturation of immature CXCR4-positive osteoblasts. Furthermore, Sox9 progenitors appear to be important in maintaining the CXCR4-positive osteoblast pool to drive osteogenesis as well as potentially regulate MES disintegration through EMT.
One of the studies focused on the soft palate, which lies posterior to the hard palate and consists of a muscular structure that also undergoes fusion (Danescu et al., 2015). Deng et al. utilized in vivo overexpression of Noggin, an antagonist of the BMP pathway, and disrupted development of muscles, tendons, and aponeuroses in the soft palate. Suppression of BMP signaling in palatal mesenchyme inhibited the differentiation of aponeurosis and tendons, which resulted in the hypoplasia of palatal muscles, along with decreased mesenchymal cell proliferation and survival due to disruption of epithelial Shh expression. Interestingly, the authors found that impaired myogenesis and tenogenesis were not involved in the clefting of the soft palate. Instead, clefting was attributed to the reduced cell proliferation and survival caused by the interrupted Shh-Gli1 signaling, and the impaired development of the aponeurosis.
Odontogenesis
Papers on tooth development focused on different aspects of cell interactions during odontogenesis. The role of cell death during tooth morphogenesis and its key effect on surrounding cell populations are reviewed by Abramyan et al. The distribution of apoptotic cells during odontogenesis is compared in different vertebrate lineages from fish to mammals. Apoptosis is introduced in light of active cellular events affecting molecular signaling and behavior in neighboring cells. Apoptosis-associated molecular signaling, including factors from intrinsic and extrinsic apoptotic pathways, as well as microRNAs and autophagic signaling are described during dental cell differentiation and morphogenesis. Apoptotic pathway disruption, alterations in apoptotic cell distribution, and dysregulation of apoptosis in mutant mouse models with mutations in non-apoptotic genes, were found to contribute to the disruption of odontogenesis.
To identify the transcriptomic characteristics of dental cells in mouse incisors, Chiba et al. performed single-cell RNA-sequencing. They focused on the identification of epithelial dental subpopulations with the aim of uncovering novel marker genes for each dental cell type that eventually contributes to enamel production. They identified two subclusters of secretory ameloblasts with a distinctive expression of Dspp and Ambn, with pseudo-time analysis finding that Dspp-positive cells differentiate into Ambn-positive ameloblasts. Their study can be used as a large resource of transcriptome data for further analyses of molecular regulation controlling the formation of enamel.
The tight junctions play a key role in ameloblast differentiation and enamel production in mouse incisors. Wang et al. examined expression patterns of claudin family members during odontogenesis and found Cldn1 and Cldn10 to be highly expressed in the dental epithelium. Cldn10 was found as a novel stratum intermedium marker and a role for Cldn10 in the regulation of dental epithelial cell differentiation during amelogenesis was confirmed by functional analysis.
The ecto-endodermal boundary in the oral cavity of axolotl was analyzed by Soukup et al. in order to determine the potential source of instructive factors that induce odontogenesis. The ecto-endodermal boundary runs through the tooth fields so that individual tooth germs display differential origins from ectodermal, endodermal, or ecto-endodermal epithelial cells. Common tooth-competent zones were found in the roof of the oral cavity and one in the mouth floor, from which five pairs of tooth fields are arranged into typical tetrapod dental patterns. By using GFP-labeled oral ectoderm, they followed the spatial relationships of early initiated teeth for individual dental fields and uncovered that the first tooth usually develops in close association to the ecto-endodermal boundary.
Eye Development
The literature on the contribution of neural crest cells to the anterior segment of the eye was reviewed by Williams and Bohnsack. They cover molecular regulation of eye development and associated congenital eye diseases, with a special focus on neural crest cell-related signaling. Rare ocular diseases such as Axenfeld-Rieger syndrome, Peters anomaly, and primary congenital glaucoma are discussed in light of key genes for neural crest cell specification or migration. Theories of disease mechanisms are also debated as an understanding of neural crest cell signaling disruption may improve the management of degenerative ocular diseases.
Disruption of eye development can also occur during the closure of the optic fissure, resulting in ocular coloboma. Cellular processes and molecular signaling, including basement membrane dynamics, cell behavior, and the fate of cells contributing to the fusion are discussed by Chan et al. Recently, a large number of model organisms spanning fish, avian, and mammalian species have been used to uncover potential avenues for new research strategies and testing of factors, which can help to prevent the occurrence of fusion defects.
Development of Branchiomeric Muscles
Head and trunk skeletal muscles have distinct functions and embryonic origins. Trunk muscles are responsible for locomotion while craniofacial muscles control eye movement, facial expression, and mastication. Cranial neural crest cells migrate to the branchial arches (BA) and along with mesoderm cells regulate the development of BA muscles (Lescroart et al., 2010).
Retinoic acid (RA) signaling is shown to be involved in the development of craniofacial muscles (Wang et al., 2019) but the mechanisms used by RA in regulating BA muscles are not well understood. Wang et al. found that RA exposure induces malformation of branchiomeric muscles in mice, this is associated with increased cell death of branchial mesodermal cells. Also, the development of hypoplastic craniofacial muscles in RA treated embryos is due to reduced expression of Pitx2, Tbx1, and MyoD, in the first and second BA. Further, over-expression of RA signaling inhibited Dlx5 and Dlx6 expression in cranial neural crest cells, which altered their communications with branchial mesoderm cells.
The neck muscles in the region connecting the head and trunk originate from the posterior BA and are associated with processes such as respiration, vocalization, swallowing, and mobility of the head (Heude et al., 2018). In the trunk, the myogenic precursor cells migrate from the ventrolateral lip of the dermomyotome to the myogenic regions within limbs, diaphragm, and tongue (Masyuk and Brand-Saberi, 2015). CXCR4/SDF-1 axis is shown to regulate the migration of muscle progenitor cells and play a crucial role in the trunk, neck, and head muscle development. Yahya et al. reviewed the roles of the CXCR4/SDF-1 axis in the development of facial and neck muscles. The key discussions include the precursors of facial expression and mastication muscles that originate from mesoderm cells of BA1 and BA2, whereas the progenitors for non-branchiomeric head muscles come from the prechordal mesoderm, and progenitor mesoderm cells of the head and somites contribute to non-somatic neck muscles. CXCR4/SDF-1 axis is involved in the migration of progenitors for the formation of branchiomeric muscles and may be responsible for muscular dystrophies.
Clinical Aspects of Disruptions of Cellular Processes
Craniofacial anomalies (CFAs) observed in 0.2% of all newborns can appear like CLP or as a syndromes involving a mutation in a single gene or chromosomal aberrations. For some CFAs, the genetic background has yet to be identified. Although CFAs can be treated with surgery, it is regularly associated with complications with feeding, speech, hearing, dentition, and healing (Hortis-Dzierbicka et al., 2012). Studies on facial development in animals and humans reveal discrepancies and often do not allow all findings in animals to be transferred to humans. To overcome this issue Parisi et al. established a living cell repository of the Cranio-/orofacial region as a clinically relevant cell model to learn CFAs, with patient-derived cells obtained from discarded tissue biopsies.
Primary cilia dysfunction or ciliopathies are often associated with craniofacial pleiotropic phenotypes. Craniofacial ciliopathic phenotypes include CLP, craniosynostosis, and micrognathia (Schock and Brugmann, 2017). Understanding the significance of the basal body is necessary to gain knowledge for therapeutic treatments because most of the genes associated with ciliopathies encode proteins specific to basal bodies, centrosomes, and centriolar satellites. Ciliogenesis is regulated by a centriolar protein encoded by the C2 domain containing 3 centriole elongation regulators (C2cd3) and mutation of this gene causes Oral-Facial-Digital syndrome (OFD) in humans. Chang et al. characterized three novel C2cd3 mouse models, a conditional knockout, and two novel CRISPR-targeted lines targeting regions in the divergent C2CD3N-C2 domain or PKC-C2 domains. Also, by examining conditional alleles with two neural crest-specific drivers, the authors showed that C2CD3N-C2 is more important for craniofacial development compared to C-terminal PKC-C2 domains.
The pinna or auricle of the ear is affected in many craniofacial syndromes including Lacrimo-auriculo-dento-digital syndrome (LADD) caused by defects in FGF signaling (Teshima et al., 2016). Zhang et al. used an FGF10 knockout mouse to elucidate the mechanisms behind the pinna defects in LADD syndrome. Loss of function of Fgf10 in mice results in smaller, cup-shaped pinnae, similar to dimorphisms seen in LADD syndrome. The phenotype is driven by increased proliferation on the outer part of the pinna together with decreased proliferation in the inner part, causing inward bending.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
MB is supported by the Czech Science Foundation (19-01205S). PG-L is supported by the Rice Creek Associate Grants, Challenge Grants SUNY Oswego, and NSF MRI (1725459). JA is supported by startup funding from the University of Michigan-Dearborn.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Marie Šulcová (LMM, IAPG, Czechia) for providing a chameleon picture. We thank all authors and reviewers for contributing to the Research Topic.
References
Barlow, A. J., and Francis-West, P. H. (1997). Ectopic Application of Recombinant BMP-2 and BMP-4 Can Change Patterning of Developing Chick Facial Primordia. Development 124, 391–398. doi:10.1242/dev.124.2.391
Buser, M. C., and Pohl, H. R. (2015). Windows of Sensitivity to Toxic Chemicals in the Development of Cleft Palates. J. Toxicol. Environ. Health B 18, 242–257. doi:10.1080/10937404.2015.1068719
Danescu, A., Mattson, M., Dool, C., Diewert, V. M., and Richman, J. M. (2015). Analysis of Human Soft Palate Morphogenesis Supports Regional Regulation of Palatal Fusion. J. Anat. 227, 474–486. doi:10.1111/joa.12365
Geetha-Loganathan, P., Nimmagadda, S., Antoni, L., Fu, K., Whiting, C. J., Francis-West, P., et al. (2009). Expression of WNT Signalling Pathway Genes during Chicken Craniofacial Development. Dev. Dyn. 238, 1150–1165. doi:10.1002/dvdy.21934
Green, R. M., Leach, C. L., Diewert, V. M., Aponte, J. D., Schmidt, E. J., Cheverud, J. M., et al. (2019). Nonlinear Gene Expression‐phenotype Relationships Contribute to Variation and Clefting in the A/WySn Mouse. Develop. Dyn. 248, 1232–1242. doi:10.1002/dvdy.110
Hallgrímsson, B., Jamniczky, H., Young, N. M., Rolian, C., Parsons, T. E., Boughner, J. C., et al. (2009). Deciphering the Palimpsest: Studying the Relationship between Morphological Integration and Phenotypic Covariation. Evol. Biol. 36, 355–376. doi:10.1007/s11692-009-9076-5
Hamburger, V., and Hamilton, H. L. (1951). A Series of normal Stages in the Development of the Chick Embryo. J. Morphol. 88 (1), 49–92. doi:10.1002/jmor.1050880104
Hammond, N. L., Dixon, J., and Dixon, M. J. (2019). Periderm: Life-Cycle and Function during Orofacial and Epidermal Development. Semin. Cel Develop. Biol. 91, 75–83. doi:10.1016/j.semcdb.2017.08.021
Heude, E., Tesarova, M., Sefton, E. M., Jullian, E., Adachi, N., Grimaldi, A., et al. (2018). Unique Morphogenetic Signatures Define Mammalian Neck Muscles and Associated Connective Tissues. Elife 7, e40179. doi:10.7554/eLife.40179
Hortis-Dzierzbicka, M., Radkowska, E., and Fudalej, P. S. (2012). Speech Outcomes in 10-Year-Old Children with Complete Unilateral Cleft Lip and Palate after One-Stage Lip and Palate Repair in the First Year of Life. J. Plast. Reconstr. Aesthet. Surg. 65, 175–181. doi:10.1016/j.bjps.2011.09.015
Hu, D., Marcucio, R. S., and Helms, J. A. (2003). A Zone of Frontonasal Ectoderm Regulates Patterning and Growth in the Face. Development 130, 1749–1758. doi:10.1242/dev.00397
Hu, D., Young, N. M., Xu, Q., Jamniczky, H., Green, R. M., Mio, W., et al. (2015). Signals from the Brain Induce Variation in Avian Facial Shape. Dev. Dyn. 244, 1133–1143. doi:10.1002/dvdy.24284
Khalil, B. D., and El-Sibai, M. (2012). Rho GTPases in Primary Brain Tumor Malignancy and Invasion. J. Neurooncol. 108, 333–339. doi:10.1007/s11060-012-0866-8
Kousa, Y. A., and Schutte, B. C. (2016). Toward an Orofacial Gene Regulatory Network. Dev. Dyn. 245, 220–232. doi:10.1002/dvdy.24341
Lescroart, F., Kelly, R. G., Le Garrec, J.-F., Meilhac, S. M., Buckingham, M., and Buckingham, M. (2010). Clonal Analysis Reveals Common Lineage Relationships between Head Muscles and Second Heart Field Derivatives in the Mouse Embryo. Development 137, 3269–3279. doi:10.1242/dev.050674
Masyuk, M., and Brand-Saberi, B. (2015). Recruitment of Skeletal Muscle Progenitors to Secondary Sites: A Role for CXCR4/SDF-1 Signalling in Skeletal Muscle Development. Results Probl. Cel Differ. 56, 1–23. doi:10.1007/978-3-662-44608-9_1
Mossey, P. A., Little, J., Munger, R. G., Dixon, M. J., and Shaw, W. C. (2009). Cleft Lip and Palate. The Lancet 374, 1773–1785. doi:10.1016/s0140-6736(09)60695-4
Richbourg, H. A., Hu, D. P., Xu, Y., Barczak, A. J., and Marcucio, R. S. (2020). miR ‐199 Family Contributes to Regulation of Sonic Hedgehog Expression during Craniofacial Development. Develop. Dyn. 249, 1062–1076. doi:10.1002/dvdy.191
Schock, E. N., and Brugmann, S. A. (2017). Discovery, Diagnosis, and Etiology of Craniofacial Ciliopathies. Cold Spring Harb Perspect. Biol. 9, a028258. doi:10.1101/cshperspect.a028258
Teshima, T. H. N., Lourenco, S. V., and Tucker, A. S. (2016). Multiple Cranial Organ Defects after Conditionally Knocking Out Fgf10 in the Neural Crest. Front. Physiol. 7, 488. doi:10.3389/fphys.2016.00488
Wang, Q., Kurosaka, H., Kikuchi, M., Nakaya, A., Trainor, P. A., and Yamashiro, T. (2019). Perturbed Development of Cranial Neural Crest Cells in Association with Reduced Sonic Hedgehog Signaling Underlies the Pathogenesis of Retinoic-Acid-Induced Cleft Palate. Dis. Model. Mech. 12, dmm040279. doi:10.1242/dmm.040279
Keywords: palatogenesis, tooth development, eye, neural crest, craniofacial
Citation: Geetha-Loganathan P, Abramyan J and Buchtová M (2022) Editorial: Cellular Mechanisms During Normal and Abnormal Craniofacial Development. Front. Cell Dev. Biol. 10:872038. doi: 10.3389/fcell.2022.872038
Received: 09 February 2022; Accepted: 18 February 2022;
Published: 08 March 2022.
Edited and Reviewed by:
Philipp Kaldis, Lund University, SwedenCopyright © 2022 Geetha-Loganathan, Abramyan and Buchtová. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Poongodi Geetha-Loganathan, p.geethaloganathan@oswego.edu
 Poongodi Geetha-Loganathan
Poongodi Geetha-Loganathan John Abramyan
John Abramyan Marcela Buchtová
Marcela Buchtová