Abstract
Increasing rates of hybridization and introgression in managed populations of freshwater fish are a major threat to the long-term viability of native species. The conservation challenge begins with identifying native gene pools. For brown trout (Salmo trutta Linnaeus, 1758) in the Upper Danube drainage, this task is complicated by the presence of both naturally and anthropogenically induced admixture of highly divergent lineages (Atlantic and Danubian). Herein, a ddRADseq protocol was used to type 377 individuals from 24 populations in the Upper Danube in Austria and Germany, and from reference populations from adjacent drainages and commercial hatcheries. High genetic differentiation at small geographic scales was found among pure Danubian-lineage populations, especially in the Kalkalpen National Park (Austria). In the Upper Danube drainage of Germany, as well as in the Rhine and Elbe drainages, brown trout populations were predominantly of Atlantic-lineage origin – as were those of all commercial hatcheries. Most populations, however, showed various degrees of admixture between Danubian and Atlantic lineages, hypothesized to be the result of both natural and anthropogenic processes. We highlight the conservation value of pure Danubian-lineage populations, and the challenges promoting conservation of naturally admixed populations, while discouraging continued stocking and admixture via management activities.
Similar content being viewed by others
Introduction
Inland fisheries management in much of continental Europe is largely private, highly fragmented in spatial terms, and characterized by a heavy reliance on stocking programs (Pinter et al., 2019; Arlinghaus et al., 2022). This is especially true for salmonid fishes, such as brown trout (Salmo trutta Linnaeus, 1758), where stocking of hatchery-reared fish or cross-basin transfers has been a common practice aimed at supporting sport fishing or artisanal interests, reaching back at least into the Middle Ages (Diem, 1964; Pechlaner, 1984; Miró & Ventura, 2013; Splendiani et al., 2016). Under such circumstances, advocating the protection of native gene pools has had limited success outside of various nature protection areas, such as national parks, or involving a few conservation-minded managers. Unfortunately, this has led to the widespread use of non-native stocking material (Weiss et al., 2001; Pinter et al., 2019; Kohout et al., 2012; Prunier et al., 2022) and thus raises serious concerns that admixture between native and non-native individuals will threaten the viability of locally adapted populations (Allendorf et al., 2001; Hansen et al., 2009).
In the Upper Danube drainage of Austria and Germany, two major mitochondrial DNA (mtDNA) lineages (Atlantic & Danubian) of brown trout occur across an extensive contact (hybrid) zone (Bernatchez et al., 1992; Riffel et al., 1995; Weiss et al., 2001; Lerceteau-Köhler et al., 2013). Recent analysis estimated that these two mtDNA lineages may share a common ancestor in the Pliocene (Veličković et al., 2023), significantly older than previously thought (Bernatchez, 2001). Nonetheless, both lineages are thought to be native to this area, with the Atlantic lineage arriving via post-glacial colonization from adjacent watersheds (Weiss et al., 2001). While the quaternary glaciations generally had major impacts on population structure, distribution, and the demographic history of freshwater fish species in Europe (Weiss & Ferrand, 2002), these natural patterns are also influenced by human-mediated translocations and subsequent admixture. Lerceteau-Köhler et al. (2013) provided the first extensive genetic analysis of 114 populations of Danubian brown trout in Austria and Germany, including nearby headwater locations in the Rhine and Elbe catchments. The results corroborated earlier findings of Schliewen et al. (2001), demonstrating that the Upper Danube in Germany is predominantly characterized by the Atlantic lineage, with little trace of diagnostic Danubian-lineage alleles. Furthermore, the hybrid zone in the Upper Danube catchment revealed a downstream gradient of an increasing proportion of Danubian ancestry. Pure Danubian-lineage populations of brown trout in the Upper Danube catchment, however, are restricted to isolated, primarily alpine, headwater streams (Baric et al., 2010; Lerceteau-Köhler et al., 2013; Schenekar et al., 2014). This is likely due to natural (i.e., ancient cross-basin colonization of Atlantic-origin brown trout) and human-mediated (i.e., large-scale, continuous stocking efforts) colonization of Atlantic-lineage brown trout into the region, which led to admixture in all but the most isolated systems. Schenekar et al. (2014) showed that hatchery and wild strains of Atlantic-lineage brown trout in the Upper Danube could be distinguished with a high-resolution microsatellite panel (17 loci) or with extensive mtDNA sequencing (> 6000 bp). Based on this study and the initial hypothesis of Weiss et al. (2001), it was inferred that admixed populations north of the Alps are predominately of natural origin but the presence of the Atlantic lineage in alpine headwater streams of the Upper Danube in Austria is primarily the result of modern supplementary stocking of Atlantic-lineage individuals. We note, however, that hatcheries in this region are primarily small, with little government oversight over their activities with respect to genetic origin of stocking material. Brood fish are routinely supplemented with wild caught fish, and there is a large amount of exchange of brood fish and offspring among hatchery owners (Pinter et al., 2019) – and stocking activities themselves are often not documented.
The genetic characterization of native stocks of brown trout throughout their natural range has generally gained considerable attention (Ryman et al., 1995; Laikre, 1999; Vera et al., 2018). This is due to both the decline in fish stocks and the need to identify locally adapted populations of brown trout for management programs. Accordingly, the distinction between natural and anthropogenic admixture of Atlantic and Danubian lineages in the Upper Danube River drainage is considered of high conservation importance, as well as the genetic characterization of native local populations. So far, population structure and interpopulation differentiation of brown trout in the Upper Danube drainage has been investigated using either exclusively mtDNA or a low number of microsatellite loci (e.g., Baric et al., 2010; Schenekar et al., 2014). Novel high throughput sequencing techniques such as double-digest restriction site associated DNA sequencing (ddRADseq) provide great possibilities for population genomic studies by drastically expanding the number of loci being analyzed (Peterson et al., 2012). Here, we apply a ddRADseq approach to brown trout populations across Austria including samples representative of the major drainages of Germany, to determine the population genetic structure, as well as signals of admixture in the Upper Danube region. Populations from the Kalkalpen National Park (Upper Austria) play a central role in this analysis, as previous investigations in this area suggested the presence of distinctive pure Danubian-lineage populations of high conservation concern.
Materials and methods
Sampling
In total, fin clips from 479 brown trout stemming from 24 locations (Fig. 1, Table 1) were sampled and stored in 95% EtOH. To serve as a reference for pure native Danubian-lineage brown trout, 56 samples were collected from three populations (GOS, ETR, RAP) known from previous studies to consist predominantly of the Danubian lineage (Lerceteau-Köhler et al., 2013). Similarly, Atlantic-lineage reference populations were sampled from four locations in Germany, totalling 77 individuals across the two main drainages: Rhine (ISN, ROW, SDA) and Elbe (SZB). Additionally, we analyzed 13 populations of the Danubian catchment north of the Alps, including samples from the Kalkalpen National Park (Upper Austria) and the Bavarian Forest National Park (Germany). Commercial stocking material of brown trout in Austria is represented by 72 individuals from four hatcheries.
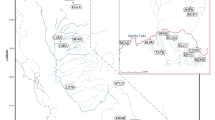
Geographic location of the study area. a geographic distribution of the two main brown trout lineages in Europe based on mtDNA data from Bernatchez (2001) and SNP data from Hashemzadeh Segherloo et al. (2021). The Ponto-Caspian lineage includes the Danubian brown trout. b Sampling sites in the Danube, Rhine and Elbe drainage systems; population codes correspond to Table 1). Base map taken from: https://en.m.wikipedia.org/wiki/File:Europe_relief_laea_location_map.jpg. c Sampling locations within the Kalkalpen National Park (national park area in green)
DNA extraction and ddRADseq libraries
Genomic DNA was extracted from fin clips using a standard Ammonium Acetate protocol (Sambrook et al., 1989). DNA integrity was assessed on a 1% Agarose gel.
The ddRADseq libraries were constructed following Peterson et al. (2012). In short, each library consisted of 48 individuals. SphI and ApoI restriction enzymes (New England Biolabs) were used to digest 500 ng of genomic DNA from each individual, for 3 h at 37 °C. The adapters (unique P1 and common P2 from Peterson et al., 2012) were ligated to the DNA fragments at 23 °C for 30 min. Thereafter, the ligated products were cleaned using 1.8X volume Ampure beads (Beckman Coulter) and pooled in equimolar proportions. The pooled samples were size-selected using a Blue Pippin (Sage Science) set to 300 bp (tight range) and amplified with a Phusion High-Fidelity PCR Kit (New England Biolabs) using PCR1 and PCR2 primers from Peterson et al. (2012). Final libraries were quantified by qPCR using the NEBNext Library Quant Kit for Illumina (New England Biolabs). Sequencing was performed by the NGS Facility at Vienna Biocenter Core Facilities (VBCF, Austria) using an Illumina HiSeq 2500 to produce 100 bp single-end reads. Illumina reads were deposited at NCBI’s Sequence Read Archive under accession numbers SAMN38751275–SAMN38751651 (Supplementary Material S1).
Raw data processing and reference genome mapping
Each HiSeq run produced on average 100 million reads, with an average of 2.1 million reads per individual (range: 27,000–9.1 million). Raw reads were demultiplexed and quality-filtered using process_radtags in Stacks v.2.55 (Catchen et al., 2011, 2013) and subsequently quality checked with MultiQC v.1.11 (Ewels et al., 2016). The 5 bp cut site was removed using Trimmomatic v.0.39 (Bolger et al., 2014), and quality filtered reads of each individual were mapped to the genome of Salmo salar Linnaeus, 1758 (GCA_000233375.4; Lien et al., 2016) using Bowtie 2 v.2.2.9 (Langmead & Salzberg, 2012). Approximately 92% of the quality filtered reads from each individual were successfully mapped to the S. salar genome. At the time of this analysis, the S. salar genome was the only well-annotated Salmo genome available and has been used in a number of studies with brown trout (e.g., Hashemzadeh Segherloo et al., 2021), despite the fact that a S. trutta genome has recently become available (GCF_901001165.1, Hansen et al., 2021). The output SAM files from Bowtie 2 analysis were sorted with samtools (Danecek et al., 2021) and used to build a catalogue of loci using the gstacks module implemented in Stacks (Catchen et al., 2011, 2013) under a minimum mapping quality (–min-mapq parameter) of 20. At this step, 42–53% of the reads were discarded due to a low mapping quality (Supplementary Material S2), which includes reads mapping multiple times and thus filtering out potential paralogs. Loci were exported using the populations program in Stacks to produce output files for downstream population genomic analyses (Catchen et al., 2011, 2013). We only retained loci present in all populations (-p parameter) and in at least 75% of the individuals in each population (-r parameter); we also discarded loci with minor allele frequencies (MAF) under 0.05 (–min-maf parameter), which can be the result of sequencing errors rather than representing true polymorphisms. In order to only analyze putatively unlinked SNPs in downstream analysis, we kept only one random SNP per locus (–write-random-snp parameter), resulting in a dataset of 2,540 SNPs. Overall, a total of 377 (78.5%) individuals across 24 populations yielded a sufficient coverage (> 5x) to be used in downstream analysis. Five LOH samples were kept despite a lower mean coverage (between 4.1–4.9x), in order to retain a minimum number of 10 individuals per population. The output file was visualized with matrix condenser (de Medeiros and Farrell, 2018; de Medeiros, 2019) in R v.4.1.2 (R Core Team, 2021) to evaluate effects of missing data by including/excluding samples with low coverage. The percentage of missing data ranged from 0.0 to 26.5% (mean 2.8%), with only one individual above 20% (Supplementary Material S2).
Detecting and retaining neutral SNPs
Certain population genetic analyses are characterized by an underlying assumption of SNP neutrality and violation of this assumption can induce significant bias in the results (Foll & Gaggiotti, 2008). Therefore, BayeScan v.2.1 (Foll & Gaggiotti, 2008) was used to detect SNPs under selection, with a posterior probability > 0.95 for outlier SNPs and prior odds set to 1, making the neutral model as likely as the model with natural selection. Such low prior odds lead to a high number of false-positives in terms of signals of selection, but it ensures that any SNP even remotely under selection is detected and removed from the dataset, thus retaining only neutral SNPs. A total of 331 SNPs were putatively under selection, leaving 2,209 neutral SNPs for downstream analyses.
Population genomic analyses
Arlequin v.3.5.2.2 (Excoffier & Lischer, 2010) was used to estimate FST values and corresponding p-values of each population, as well as to evaluate the deviation from Hardy Weinberg Equilibrium (HWE) for each population by computing the inbreeding coefficients (FIS values), expected heterozygosity (HE) and observed heterozygosity (HO) with an allowed missing level of 0.1.
Clustering analyses
To help evaluate the population structure of brown trout in the Upper Danube drainage and visualize inter-cluster differentiation with no prior assumptions related to genetic models, a Principal Components Analyses (PCA) was carried out on SNP genotypes. The analysis was performed using the R packages adegenet v.2.1.8 (Jombart, 2008; Jombart & Ahmed, 2011) and factoextra v.1.0.7 (Kassambara & Mundt, 2020) in R v.4.1.2.
The population structure and shared ancestry were also investigated using the program ADMIXTURE v.1.3.0 (Alexander et al., 2009), which estimates individual ancestry from multilocus SNP datasets using maximum likelihood in a parametric model. Varying numbers of assumed population clusters (K) were tested ranging from 1 to 24, each with 10 independent runs. The results of all replicates for each K were summarized using the program CLUMPAK (Kopelman et al., 2015) in StructureSelector (Li & Liu, 2018). A separate ADMIXTURE analysis was run for Atlantic reference-, hatchery- and admixed Danubian populations (based on 2,291 neutral SNPs). The optimal number of clusters was evaluated based on the cross-validation error.
Phylogenetic structure
A maximum likelihood (ML) approach implemented in IQtree v.2.1.4 (Minh et al., 2020) was used to roughly depict the genetic relationships among brown trout in the Upper Danube and adjacent drainages. The best-fit nucleotide substitution model (TVM + F + R8) was identified using ModelFinder (Kalyaanamoorthy et al., 2017) implemented in IQtree with three independent runs and based on the Bayesian Information Criterion (BIC). A concatenated alignment of all ddRADseq loci was used to infer a ML tree with 1,000 ultrafast bootstrap replicates and applying the -bnni flag to reduce the impact of overestimating branch support with ultrafast bootstraps (Hoang et al., 2018).
Results
Genetic differentiation
Expected heterozygosity (HE) ranged from 0.146 (RMB, ETR) to 0.364 (BRB) and was generally lower than observed heterozygosity (HO) with the exception of populations AND, LOH, and ETR (Table 2). Most FIS values (inbreeding coefficient) were negative, indicating heterozygosity excess in the populations. This is expected with genomic markers and very small population sizes, which most of our wild samples from headwater streams represent. Based on the FIS values, 10 of 24 populations showed a significant deviation from Hardy–Weinberg equilibrium (at P < 0.05), however after applying a table-wide sequential Bonferroni correction (Sokal & Rohlf, 1995), only four populations (JGB, SAB, KSL, NIB) remained statistically significant. Pairwise FST values, measuring differentiation among populations, were lowest among populations from the Krumme Steyrling catchment (KSL, RMB, SGB; FST = 0.004–0.060), and among populations from the Rhine drainage (SDA, ISN, ROW; FST = 0.077–0.177) (Supplementary Material S3). The highest FST values were found between Atlantic and Danubian-lineage populations, ranging from 0.666 (SDA vs. SAB) to 0.827 (ISN vs. GOS) reflecting the deep divergence between these major phylogeographic lineages. Within the Upper Danube drainage, the highest FST value was found between the northern Alpine SEE and GOS (0.721) populations; FST values among populations from the Kalkalpen National Park ranged from 0.004 (SGB vs. KSL) to 0.645 (SGB vs. SIB).
Population structure
In the PCA, the first axis (34.4% of variance) clearly revealed differentiation between pure native Danubian populations (ETR, GOS, RAP) as well as some populations from the Kalkalpen National Park (NIB, SAB, STG, KSL, RMB, SGB) to Atlantic lineage reference populations (ISN, SZB, SDA, ROW) (Fig. 2a). Samples from the Upper Elbe grouped together with those from the Rhine. Other Danubian populations, including samples from hatcheries (HTL, AND, WES, THA), appeared to be admixed with strong Atlantic influence. The Danubian SEE population from the Bavarian Forest National Park grouped together with the Atlantic reference populations. Variability among Danubian-lineage populations was observed along PC2 (3.3% of variance) and PC3 (2.6% of variance), where samples from the Kalkalpen National Park were distinct from Danubian reference populations and displayed differentiation among distinct drainage systems at small geographic scales. Strong within population variation was not only observed for hatchery samples but was also present in wild (admixed and reference) populations; e.g., ETR, which appeared as a pure Danubian population, with exception of one individual (ETR120) which seemed to be of admixed origin, grouping with individuals of BRB, JGB, LOH.
Population structure and phylogenetic relationships of the 24 analyzed brown trout populations. a Principal component analysis (PC1 vs. PC2) and (PC1 vs. PC3). Each circle represents one individual and circles are color coded for population origin. b ADMIXTURE analysis showing clustering of individuals based on shared ancestry, each bar represents one individual. Optimal number of clusters based on the cross-validation error K = 2 or K = 17 (entire dataset); and K = 2 or K = 9 (partial dataset). c Maximum likelihood tree representing phylogenetic relationships
The ADMIXTURE analysis revealed considerable complexity among the populations analyzed but showed that the most probable number of clusters was K = 2 (strongest decrease of the cross-validation error, CV error = 0.388) or K = 17 (lowest cross validation error, CV error = 0.301), reflecting the distinction between Atlantic and Danubian lineages but also strong drainage specific sub-structure respectively (Fig. 2b; Supplementary Material S4, showing K = 2 to K = 17). Several populations in the Danube drainage north of the Alps, including the Kalkalpen National Park (SIB, JGB), appeared to be admixed. The hatchery samples were not homogenous with a distinction between AND/HTL and THA/WES at K = 17. Samples from LOH, a population previously considered to be of pure Danubian origin (Schenekar et al., 2014), was clearly admixed with only one individual (LOH304) showing a high ancestry proportion with the Danubian lineage. To better evaluate the introgression of hatchery lineages into admixed populations, we re-ran ADMIXTURE without pure Danubian populations (Fig. 2b; Supplementary Material S5, showing K = 2 to K = 9). At K = 2 (strongest decrease of the cross-validation error, CV error = 0.437) it was clear that hatchery populations appeared admixed compared to Atlantic reference populations, and at K = 9 (lowest cross validation error, CV error = 0.388) the Atlantic reference population were distinct and most similar to the Danubian SEE population, but no direct association with these potential sources, as well as hatchery populations (excluding specific individuals) was seen in the admixed populations.
Phylogenetic structure
Reference samples of the Atlantic and Danubian lineages were recovered as distinct groups (Fig. 2c) with strong support in ML analysis (bootstrap values, bs 99 and 95, respectively). Samples from the Danubian SEE population formed a well-supported (bs 99) sister group to samples from the Vltava River system (SZB). The placement of admixed individuals/populations was only weakly supported but they appeared more associated with the Atlantic lineage, including the hatcheries. Samples from the Kalkalpen National Park were not homogenous. Most of them grouped with Danubian reference populations (KSL, RMB, SGB, NIB, STG, SAB), forming well-supported subgroups (bs 95–100) reflecting their origin from distinct drainage systems. Populations JGB and SIB grouped with samples of predominantly Atlantic-lineage background, similar to the Lower Austrian LOH population where one sample (LOH304) was associated with individuals from the Krumme Steyrling (KSL, RMB, SGB).
Discussion
This study provides the first insights into the genetic structure of brown trout in the Upper Danube drainage using genome-wide SNP markers. The results 1) support significant genetic differentiation among native Danubian-lineage populations in Austria, 2) highlight the discovery of the densest cluster of pure native Danubian-lineage brown trout populations thus far found in the region (Kalkalpen National Park), and 3) support previous studies suggesting a complex evolutionary history of brown trout in the Upper Danube likely the result of both natural and anthropogenic processes. Collectively, these results present a challenge to both conservation policy and communication with stake holders responsible for fisheries management.
Previous studies have inferred an extensive contact zone of the Atlantic & Danubian lineages of brown trout in the Upper Danube drainage of Germany and Austria with a downstream gradient of increasing proportion of Danubian ancestry (Weiss et al., 2001; Lerceteau-Köhler et al., 2013). While pure Danubian-lineage brown trout in Germany have not yet been found, a few such populations have been described in Austria north of the Alps, primarily restricted to isolated headwater reaches of streams in proximity to late Pleistocene glacial margins (Baric et al., 2010; Lerceteau-Köhler et al., 2013). The significant genetic differentiation found among these native Danubian populations (Fig. 2) likely reflects high levels of genetic drift acting on such small populations with low effective population sizes (Frankham, 2005; Whitely et al., 2018) promoting isolation-by-distance (Toczydlowski & Waller, 2018), or specifically, isolation-by-resistance (sensu McRae, 2006) as dispersal is restricted. Alternatively, post-glacial expansion of brown trout in this region may have originated from multiple local glacial refugia, reflecting complex biogeographic patterns in other cold-stenothermic aquatic taxa (Malicky, 2006; Graf et al., 2014). For brown trout and other rheophilic fish species, it may thus be plausible that naturally isolated headwater populations exist along the entire Alpine range (see Fumagilli et al., 2002; Gil et al., 2016; Polgar et al., 2022) where headwater isolation of such relict populations was enhanced by erosional processes creating barriers (i.e., waterfalls) to upstream fish migration.
One such example may be found in the Kalkalpen National Park (Upper Austria). This area is located in the eastern-most extension of the Upper Austrian Prealps, which was situated just outside of the main Alpine glacial sheet during Last Glacial Maximum (van Husen, 1971, 1975). The brown trout populations in the Kalkalpen National Park are unique as they represent the densest cluster of pure Danubian-lineage brown trout found north of the Alps so far (compare to Lerceteau-Köhler et al., 2013), while showing clear genetic differentiation across small spatial scales (Fig. 2, Table 2). Most of these populations inhabit headwater creeks at 750–1090 m above sea level, in an area less than 200km2, and most of them are separated from downstream reaches by cascades. Despite the unique genetic structure observed among populations in the Kalkalpen National Park, there were clear signs of admixture in some populations (e.g., SIB, JGB, SAB) with the presence of non-Danubian alleles possibly stemming from stocking of hatchery-reared strains. Both the JGB and the SIB populations revealed Atlantic-lineage mtDNA or a high frequency of the LDH-90 allele (Weiss et al., 2017), which is common among Atlantic-lineage hatchery stocks in the region (Lerceteau-Köhler et al., 2013) and used as a diagnostic marker for Atlantic-lineage populations across central and northern Europe (Hamilton et al., 1989; Riffel et al., 1995). While such admixture apparently affects populations in the downstream reaches (JGB, SIB) more than those in the headwaters, it may still pose a potential threat toward sustaining the genetic integrity of native brown trout populations in the region. Conservation efforts of the Kalkalpen National Park have already targeted the protection of these populations, especially in the more isolated headwaters, where, for example, introduced rainbow trout (Oncorhynchus mykiss (Walbaum, 1792)) has been eliminated after an intensive ten-year-long removal campaign (Weiss & Schenekar, 2021).
Besides the detection of pure native Danubian-lineage populations, the newly obtained genome wide SNP data offer a more detailed view on formally identified pure Danubian-lineage populations. Schenekar et al. (2014) reported such a population outside the Alpine reach, located in a small and isolated tributary creek (2–3 m wide) of the Kamp River drainage (Lohnbach, LOH). The SNP-based analysis, however, clearly demonstrated that this population is of admixed origin, despite the fixation of both Danubian-lineage mtDNA and the ancestral LDH-100 allele (Hamilton et al., 1989; Schenekar et al., 2014), which until now was only found to be fixed in pure Danubian populations (Lerceteau-Köhler et al., 2013). The fixation of Danubian mtDNA or the LDH-100 allele could perhaps occur in such a small population through genetic drift (Raeymaekers et al., 2008; Carrara et al., 2014). While the population is clearly admixed, the fact that a typical hatchery-reared genetic profile (i.e., Atlantic-lineage mtDNA or the LDH-90 allele) is missing, may support that this admixture is more ancient and potentially natural. A natural cause of admixture in LOH seems only plausible via the Upper Danube drainage, since a direct connection from the Kamp River drainage to the Atlantic basin (Elbe headwaters) seems unlikely since the Pleistocene (Fischer, 1979). Such natural admixture is concordant with the original hypothesis of Weiss et al. (2001), reiterated with a larger data set, and analyses in Lerceteau-Köhler et al. (2013). However, the origin (natural or anthropogenic) of the admixture in the LOH population remains conjectural at this point, but Casanova et al. (2022) point out that genomic SNP markers are better suited in detecting introgression of Atlantic-lineage fish, because the bi-allelic LDH-C locus can reveal high percentages of an LDH-100 homozygote, even in F2 generation crosses, between local native and hatchery-reared strains. More broadly, it is noteworthy that admixed populations do not show a general pattern of association with either the Atlantic reference populations used in this study, nor the Atlantic lineage hatchery populations, beyond the potential recognition of a few individual fish, which likely reflect either hatchery fish, or recently introgressed fish (Fig. 2b). In our view, this is most easily explained by more ancient (i.e., natural) admixture with unknown source populations. While additional approaches have been applied for attempting to better differentiate between more ancient and recent introgression in Salmo sp. using SNPs, these studies are in different geographic regions where the Atlantic lineage of brown trout is exotic with no chance of natural occurrence, and either apply much more dense arrays of markers or a pre-screened set of markers tested for diagnostic power (Sušnik Bajec et al., 2015; Leitwein et al., 2018; Saint-Pé et al., 2019). Our lack of information on hatchery brood stocks, which are not kept isolated from wild supplementation, our limited number of markers, and most of all clear understanding that the Atlantic-lineage of brown trout is also natural in the region limits our ability to better address this question with the given set of SNPs.
Overall, the post-glacial colonization routes of freshwater fishes in the Upper Danube region are poorly understood and for brown trout it remains a mystery how the upper Danubian drainage (i.e., in Bavaria) became overwhelmingly dominated by the Atlantic lineage (Schliewen et al., 2001; Lerceteau-Köhler et al., 2013). Noteworthy in this respect, is the Atlantic lineage SEE population in the Bavarian Forest National Park. Another important point is that typical hatchery material in this region does not genetically group closely with alleged natural Atlantic lineage populations, whether found in the Rhine, Elbe, or upper Danube River drainages (Fig. 2b, but see also Lerceteau-Köhler et al., 2013). This likely stems from the fact that many Atlantic-lineage hatchery strains share a common ancestry, supported by commercial networking and stock exchange (Pinter et al., 2017), which is not locally restricted, but largely similar throughout much of Central- and Western Europe (Sanz et al., 2020; Berrebi et al., 2021; Casanova et al., 2022). Thus, as shown by Casanova et al. (2022), it is both possible and desirable to distinguish between natural and anthropogenic introgression for conservation management goals.
Conservation and management implications
For the Upper Danube region, a more systematic survey of populations with genome-wide markers, such as those presented here, coupled with a diagnostic approach (e.g., Casanova et al., 2022), will be necessary to provide a better overall picture of the distribution and distinction of allegedly naturally vs. anthropogenically induced admixed populations with respect to the Danubian and Atlantic lineages. Such knowledge may contribute to more general or regional recommendations of how to best manage populations of brown trout in the Upper Danube River drainage as well as adjacent catchments. For the time being it has become clear that the drainage-bearing epithets “Danubian” and “Atlantic” lineage, presented in peer-reviewed literature for over 30 years, are not suitable to identify native gene pools of brown trout in the region, nor do they necessarily help in assigning a conservation status to specific populations.
Nonetheless, pure Danubian-lineage populations thus far identified are exceedingly rare, and as such should be assigned a high conservation status in the region. In recent years, a small market demand has arisen for native Danubian trout, although very few such stocks exist, and thus availability is limited. The results of this study underscore that among the few available populations of pure native Danubian lineage brown trout, significant genetic variation exists even across small geographic distances. Using any such population as a source of brood fish for widespread stocking runs counter to conservation genetic principles that support the assumption that each sub-population may contain local adaptations or co-adapted gene complexes that might benefit populations in their native habitat but not necessarily anywhere else (Allendorf et al., 2001; Hansen et al., 2009; Prunier et al., 2022).
Another concern is that as fishery managers become increasingly familiar with the fact that most populations in the region are admixed, inhibition to stocking rivers with foreign strains of trout might be eroded. Here it is important to communicate the clear difference between naturally vs. anthropogenically induced admixture. Allendorf et al. (2001) recognized six categories of hybridization and introgression in a conservation context, concluding that three of these categories encompass natural hybridization and introgression (whether inter- or intraspecific) and are a normal part of the evolutionary process and thus do not pose a conservation concern. On the other hand, anthropogenically induced hybridization and introgression, especially involving hatchery-reared individuals is considered problematic because it promotes the spread of mal-adaptive genetic variation (Bolstad et al., 2017, 2021; Besnier et al., 2022). For naturally admixed populations, especially stemming from contact events or past hybridization dating back to several thousand years ago, natural selection can steer adaptation and fitness most likely at a pace amenable to the biological and environmental limitations of the system. For reared salmonid fishes, however, artificial selection within the hatcheries almost invariably results in lower survival and fitness decline, a process that can already begin after a single generation in captivity (Araki et al., 2007; Fraser et al., 2019). Thus, hatchery-rearing and stocking cannot be endorsed independent of whether or not the genetic background of the fish belongs to a major lineage that may be perceived as native in a particular drainage. Furthermore, populations with natural patterns of ancient admixture should be provided the same conservation status as non-admixed populations (Allendorf et al., 2001), although some priorities may be established for rare populations, for example, for pure Danubian-lineage brown trout vs. the more widespread occurrence of admixed populations.
In light of the market-driven forces underlying most fishery management plans and decisions in the region, the value of protected areas, such as the Kalkalpen National Park in Austria, or the Bavarian Forest National Park in Germany where no such market pressure is present, cannot be overstated. River systems in these areas may be among the very last in the Upper Danube catchment, where conservation has explicit legislative support and thus evolutionary processes can proceed in a natural way, without disruption from the introduction of non-native or hatchery-reared strains of fish.
Data availability
Raw ddRADseq reads were deposited at NCBI’s Sequence Read Archive (BioProject: PRJNA1050776) under accession numbers SAMN38751275–SAMN38751651 (Supplementary Material S1).
References
Alexander, D. H., J. Novembre & K. Lange, 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Research 19: 1655–1664. https://doi.org/10.1101/gr.094052.109.
Allendorf, F. W., R. F. Learya, P. Spruella & J. K. Wenburg, 2001. The problems with hybrids: setting conservation guidelines. Trends in Ecology & Evolution 16: 613–622. https://doi.org/10.1016/S0169-5347(01)02290-X.
Araki, H., B. Cooper & M. S. Blouin, 2007. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science 318: 100–103. https://doi.org/10.1126/science.1145621.
Arlinghaus, R., C. Riep, S. Theis, T. Pagel & M. Fujitani, 2022. Dysfunctional information feedbacks cause the emergence of management panaceas in social-ecological systems: the case of fish stocking in inland recreational fisheries. Journal of Outdoor Recreation and Tourism 38: 100475. https://doi.org/10.1016/j.jort.2021.100475.
Baric, S., A. Riedl, A. Meraner, N. Medgyesy, R. Lackner, B. Pelster & J. Dalla Via, 2010. Alpine headwater streams as reservoirs of remnant populations of the Danubian clade of brown trout. Freshwater Biology 55: 866–880. https://doi.org/10.1111/j.1365-2427.2009.02318.x.
Bernatchez, L., 2001. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55: 351–379. https://doi.org/10.1111/j.0014-3820.2001.tb01300.x.
Bernatchez, L., R. Guyomard & L. Bonhomme, 1992. DNA sequence variation of the mitochondrial control region among geographically and morphologically remote European brown trout Salmo trutta populations. Molecular Ecology 1: 161–173. https://doi.org/10.1111/j.1365-294X.1992.tb00172.x.
Berrebi, P., A. Horvath, A. Splendiani, S. Palm & R. Bernás, 2021. Genetic diversity of domestic brown trout stocks in Europe. Aquaculture 544: 737043.
Besnier, F., F. Ayllon, O. Skaala, M. F. Solberg, P. T. Fjeldheim & K. Anderson, 2022. Introgression of domesticated salmon changes life history and phenology of a wild salmon population. Evolutionary Applications 15: 853–864. https://doi.org/10.1111/eva.13375.
Bolger, A. M., M. Lohse & B. Usadel, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. https://doi.org/10.1093/bioinformatics/btu170.
Bolstad, G. H., K. Hindar, G. Robertsen, B. Jonsson, H. Sægrov, O. H. Diserud, P. Fiske, A. J. Jensen, K. Urdal, T. F. Næsje, B. T. Barlaup, B. Florø-Larsen, H. Lo, E. Niemelä & S. Karlsson, 2017. Gene flow from domesticated escapes alters the life history of wild Atlantic salmon. Nature Ecology & Evolution 1: 0124. https://doi.org/10.1038/s41559-017-0124.
Bolstad, G. H., L. Karlsson, I. J. Hagen, P. Fiske, K. Urdal, H. Sægrov, B. Florø-Larsen, V. P. Sollien, G. Østborg, O. H. Diserud, A. J. Jensen & K. Hindar, 2021. Introgression from farmed escapees affects the full life cycle of wild Atlantic salmon. Science Advances 7: eabj3397. https://doi.org/10.1126/sciadv.abj3397.
Carrara, F., A. Rinaldo, A. Giometto & F. Altermatt, 2014. Complex interaction of dendritic connectivity and hierarchical patch size on biodiversity in river-like landscapes. The American Naturalist 183: 13–25.
Casanova, A., S. Heras, A. Abras, M. I. Roldán, C. Bouza, M. Vera, J. L. García-Marin & P. Martínez, 2022. Genomic hatchery introgression in brown trout (Salmo trutta L.): development of a diaganostic SNP Panel for monitoring the impacted Mediterranean rivers. Genes 13: 255. https://doi.org/10.3390/genes13020255.
Catchen, J. M., A. Amores, P. Hohenlohe, W. Cresko & J. H. Postlethwait, 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3: Genes Genomes, Genetics 1: 171–182. https://doi.org/10.1534/g3.111.000240.
Catchen, J., P. Hohenlohe, S. Bassham, A. Amores & W. Cresko, 2013. Stacks: an analysis tool set for population genomics. Molecular Ecology 22: 3124–3140. https://doi.org/10.1111/mec.12354.
Danecek, P., J. K. Bonfield, J. Liddle, J. Marshall, V. Ohan, M. O. Pollardet, A. Whitwham, T. Keane, S. A. McCarthy, R. M. Davies & H. Li, 2021. Twelve years of SAMtools and BCFtools. GigaScience 10: giab008.
de Medeiros, B. A. S. & B. D. Farrell, 2018. Whole-genome amplification in double-digest RADseq results in adequate libraries but fewer sequenced loci. PeerJ 6: e5089. https://doi.org/10.7717/peerj.5089.
Diem, H., 1964. Beiträge zur Fischerei Nordtirols, Teil B, Die Fischerei in den natürlichen Gewässern der Vergangenheit. Veröffentlichungen Des Musesums Ferdinandeum 43: 5–132 ([In German]).
Ewels, P., M. Magnusson, S. Lundin & M. Käller, 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. https://doi.org/10.1093/bioinformatics/btw354.
Excoffier, L. & H. E. L. Lischer, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x.
Fischer, H., 1979. Reliefgenerationen im Kristallinmassiv Donauraum, Alpenvorland und Alpenrand im westl. Niederösterreich. Forschungen zur deutschen Landeskunde 213: 232 pp. [in German]
Foll, M. & O. E. Gaggiotti, 2008. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 180: 977–993. https://doi.org/10.1534/genetics.108.092221.
Frankham, R., 2005. Genetics and extinction. Biological Conservation 126: 131–140. https://doi.org/10.1016/j.biocon.2005.05.002.
Fraser, D. J., L. Walker, M. C. Yates, K. Marin, J. L. A. Wood, T. A. Bernos & C. Zastavniouk, 2019. Population correlates of rapid captive-induced maladaptation in a wild fish. Evolutionary Applications 12: 1305–1317. https://doi.org/10.1111/eva.12649.
Fumagilli, L., A. Snoj, D. Jesensek, F. Balloux, T. Jug, O. Duron, F. Brossier, A. J. Crivelli & P. Berrebi, 2002. Extreme genetic differentiation among the remnant populations of marble trout (Salmo marmoratus) in Slovenia. Molecular Ecology 11: 2711–2716. https://doi.org/10.1046/j.1365-294X.2002.01648.x.
Gil, J., J. Labonne & A. Caudron, 2016. Evaluation of strategies to conserve and restore intraspecific biodiversity of brown trout: outcomes from genetic monitoring in the French Alps. Reviews in Fish Biology and Fisheries 26: 1–11. https://doi.org/10.1007/s11160-015-9405-y.
Graf, W., M. Konar, D. Murányi, K. Orci & S. Vitecek, 2014. A new species of Isoperla (Insecta, Plecoptera) from the Karawanken, with considerations on the Southern Limestone Alps as centers of endemism. ZooKeys 448: 27–36. https://doi.org/10.3897/zookeys.448.8509.
Hamilton, K. E., A. Ferguson, J. B. Taggart, T. Tómasson, A. Walker & E. Fahy, 1989. Post-glacial colonization of brown trout, Salmo trutta L.: Ldh-5 as a phylogeographic marker locus. Journal of Fish Biology 35: 651–664. https://doi.org/10.1111/j.1095-8649.1989.tb03017.x.
Hansen, M. M., D. J. Fraser, K. Meier & K.-L.D. Mensberg, 2009. Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Molecular Ecology 178: 2549–2562. https://doi.org/10.1111/j.1365-294X.2009.04198.x.
Hansen, T., P. G. Fjelldal, S. Lien, M. Smith, C. Corton, K. Oliver, J. Skelton, E. Betteridge, J. Doulcan, O. Fedrigo, J. Mountcastle, E. Jarvis, S. A. McCarthy, W. Chow, K. Howe, J. Torrance, J. Wood, Y. Sims, L. Haggerty, R. Challis, J. Threlfall, D. Mead, R. Durbin & M. Blaxter, 2021. The genome sequence of the brown trout, Salmo trutta Linnaeus 1758 [version 1; peer review: 3 approved]. Wellcome Open Research 6: 108–110. https://doi.org/10.12688/wellcomeopenres.16838.1.
Hashemzadeh Segherloo, I., J. Freyhof, P. Berrebi, A.-L. Ferchaud, M. Geiger, J. Laroche, B. A. Levin, E. Normandeau & L. Bernatchez, 2021. A genomic perspective on an old question: Salmo trouts or Salmo trutta (Teleostei: Salmonidae)? Molecular Phylogenetics and Evolution 162: 107204. https://doi.org/10.1016/j.ympev.2021.107204.
Hoang, D. T., O. Chernomor, A. von Haeseler, B. Q. Minh & L. S. Vinh, 2018. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281.
Jombart, T., 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. https://doi.org/10.1093/bioinformatics/btn129.
Jombart, T. & I. Ahmed, 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. https://doi.org/10.1093/bioinformatics/btr52.
Kalyaanamoorthy, S., B. Q. Minh, T. K. F. Wong, A. von Haeseler & L. S. Jermiin, 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/NMETH.4285.
Kassambara, A. & F. Mundt, 2020. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. https://CRAN.R-project.org/package=factoextra.
Kohout, J., I. Jašková, I. Papoušek, A. Šedivá & V. Šlechta, 2012. Effects of stocking on the genetic structure of brown trout Salmo trutta in Central Europe inferred from mitochondrial and nuclear DNA markers. Fisheries Management and Ecology 19: 252–263. https://doi.org/10.1111/j.1365-2400.2011.00828.x.
Kopelman, N. M., J. Mayzel, M. Jakobsson, N. A. Rosenberg & I. Mayrose, 2015. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources 15: 1179–1191. https://doi.org/10.1111/1755-0998.12387.
Laikre, L. (ed.), 1999. Conservation genetic management of brown trout (Salmo trutta) in Europe. Report by the concerted action on identification, management and exploitation of genetic resources in the Brown trout (Salmo trutta) (“Troutconcert”; EU FAIR CT97-3882).
Langmead, B. & S. Salzberg, 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359. https://doi.org/10.1038/nmeth.1923.
Leitwein, M., P.-A. Gagnaire, E. Desmarais, P. Berrebi & B. Guinard, 2018. Genomic consequences of a recent three-way admixture in supplemented wild brown trout populations revealed by local ancestry tracts. Molecular Ecology 27: 3466–3483.
Lerceteau-Köhler, E., U. Schliewen, T. Kopun & S. Weiss, 2013. Genetic variation in brown trout Salmo trutta across the Danube, Rhine, and Elbe headwaters: a failure of the phylogeographic paradigm? BMC Evolutionary Biology 13: 176.
Li, Y. L. & J. X. Liu, 2018. StructureSelector: a web-based software to select and visualize the optimal number of clusters using multiple methods. Molecular Ecology Resources 18: 176–177. https://doi.org/10.1111/1755-0998.12719.
Lien, S., B. F. Koop, S. R. Sandve, J. R. Miller, M. P. Kent, T. Nome, T. R. Hvidsten, J. S. Leong, D. R. Minkley, A. Zimin, F. Grammes, H. Grove, A. Gjuvsland, B. Walenz, R. A. Hermansen, K. von Schalburg, E. B. Rondeau, A. Di Genova, J. K. A. Samy, J. O. Vik, M. D. Vigeland, L. Caler, U. Grimholt, S. Jentoft, D. I. Våge, P. de Jong, T. Moen, M. Baranski, Y. Palti, D. R. Smith, J. A. Yorke, A. J. Nederbragt, A. Tooming-Klunderud, K. S. Jakobsen, X. Jiang, D. Fan, Y. Hu, D. A. Liberles, R. Vidal, P. Iturra, S. J. M. Jones, I. Jonassen, A. Maass, S. W. Omholt & W. S. Davidson, 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533: 200–205. https://doi.org/10.1038/nature17164.
Malicky, H., 2006. Mitteleuropäische (extra - mediterrane) Arealkerne des Dinodal am Beispiel von Köcherfliegen (Trichoptera). Beiträge Zur Entomologie 56: 347–359 ([in German]).
McRae, B. H., 2006. Isolation by resistance. Evolution 60: 1551–1561. https://doi.org/10.1111/j.0014-3820.2006.tb00500.x.
de Medeiros, B. A., 2019. Matrix Condenser v.1.0. https://github.com/brunoasm/matrix_condenser/
Minh, B. Q., H. A. Schmidt, O. Chernomor, D. Schrempf, M. D. Woodhams, A. von Haeseler & R. Lanfear, 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534.
Miró, A. & M. Ventura, 2013. Historical use, fishing management and lake characteristics explain the presence of non-native trout in Pyrenean lakes: implications for conservation. Biological Conservation 167: 17–24.
Pechlaner, R., 1984. Historical Evidence for the Introduction of Arctic Charr into High-mountain Lakes of the Alps by Man. In Johnson, J. & B. L. Burns (eds), Biology of the Arctic Charr. Proceedings of the International Symposium on Arctic Charr University of Manitoba Press, Winnipeg: 449–557.
Peterson, B. K., J. N. Weber, E. H. Kay, H. S. Fisher & H. E. Hoekstra, 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7: e37135. https://doi.org/10.1371/journal.pone.0037135.
Pinter, K., J. Epifanio & G. Unfer, 2019. Release of hatchery-reared brown trout (Salmo trutta) as a threat to wild populations? A case study from Austria. Fisheries Research 219: 105296. https://doi.org/10.1016/j.fishres.2019.05.013.
Pinter, K., G. Unfer, B. Lundsgaard-Hansen & S. Weiss, 2017. Besatzwirtschaft in Österreich und mögliche Effekte auf die innerartliche Vielfalt der Bachforellen. Österreichs Fischerei 1: 70. Jahrgang, 15–33 [in German]
Polgar, G., M. Iaia, T. Righi & P. Volta, 2022. The Italian alpine and subalpine trouts: taxonomy, evolution, conservation. Biology 11: 576. https://doi.org/10.3390/biology11040576.
Prunier, J. G., K. Saint-Pé, L. Tissot, N. Poulet, G. Marselli, C. Veyssiére & S. Blanchet, 2022. Captive-bred ancestry affects spatial patterns of genetic diversity and differentiation in brown trout (Salmo trutta) populations. Aquatic Conservation of Marine and Freshwater Ecosystems 32: 1529–1543. https://doi.org/10.1002/aqc.3826.
R Core Team, 2021. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org.
Raeymaekers, J. A. M., G. E. Maes, S. Geldof, I. Hontis, K. Nackaerts & F. A. M. Volckaert, 2008. Modelling genetic connectivity in sticklebacks as a guideline for river restoration. Evolutionary Applications 1: 475–488. https://doi.org/10.1111/j.1752-4571.2008.00019.x.
Riffel, M., V. Storch & A. Schreiber, 1995. Allozyme variability of brown trout (Salmo trutta L.) populations across the Rhenanian-Danubian watershed in southwest Germany. Heredity 74: 241–249. https://doi.org/10.1038/hdy.1995.37.
Ryman, N., F. Utter & L. Laikre, 1995. Protection of intraspecific biodiversity of exploited fishes. Reviews in Fish Biology and Fisheries 5: 417–446. https://doi.org/10.1007/BF01103814.
Saint-Pé, K., M. Leitwein, L. Tissot, N. Poulet, B. Buinard, P. Berrebi, G. Marselli, J.-M. Lascaux, P.-A. Gagnaire & S. Blanchet, 2019. Development of a large SNPs resource and a low-density array for brown trout (Salmo trutta) population genetics. BMC Genomics 20: 582. https://doi.org/10.1186/s12864-019-5958-9.
Sambrook, J., E. Fritsch & T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Cold Harbor Spring Press, New York:
Sanz, N., A. Nebot & R.-M. Araguas, 2020. Microsatellites as a good approach for detecting triploidy in brown trout hatchery stocks. Aquaculture 523: 735218.
Schenekar, T., E. Lerceteau-Köhler & S. Weiss, 2014. Fine-scale phylogeographic contact zone in Austrian brown trout Salmo trutta reveals multiple waves of post-glacial colonization and a pre-dominance of natural versus anthropogenic admixture. Conservation Genetics 15: 561–572. https://doi.org/10.1007/s10592-013-0561-0.
Schliewen, U., C. Engelbrecht, K. Rassmann, M. Miller, L. Klein & D. Tautz, 2001. Veränderungen der genetischen Vielfalt: Molekulare und populationsökologische Charakterisierung autochthoner und durch Besatz beeinflusster Salmonidenpopulationen in Bayern. Schriftenreihe Texte des Umweltbundesamtes 48/01, Berlin [in German].
Sokal, R. R. & F. J. Rohlf, 1995. Biometry – The Principles and Practice of Statistics in Biological Research, 3rd ed. W. H. Freeman and Co, NY: 887.
Splendiani, A., P. Ruggeri, M. Giovannotti, S. Pesaresi, G. Occhipinti, T. Fioravanti, M. Lorenzoni, P. Nisi Cerioni & V. Caputo Barucchi, 2016. Alien brown trout invasion of the Italian peninsula: the role of geological, climate and anthropogenic factors. Biological Invasions 18: 2029–2044.
Sušnik Bajec, S., G. Pustovrh, D. Jesenšek & A. Snoj, 2015. Population genetic SNP analysis of marble and brown trout in a hybridization zone of the Adriatic watershed in Slovenia. Biological Conservation 184: 239–250.
Toczydlowski, R. H. & D. M. Waller, 2018. Drift happens: Molecular genetic diversity and differentiation among populations of jewelweed (Impatiens capensis Meerb.) reflect fragmentation of floodplain forests. Molecular Ecology 28: 2459–2475.
van Husen, D., 1971. Zum Quartär des unteren Ennstales von Großraming bis zur Donau. Verhandlungen Der Geologischen Bundesanstalt 3: 511–521 ([in German]).
van Husen, D., 1975. Die quartäre Entwicklung des Steyrtales und seiner Nebentäler. Jahrbuch Des Oberösterreichischen Musealvereines 120: 271–289 ([in German]).
Veličković, T., A. Snoj, V. Simić, R. Šanda, J. Vukić, D. Barcyté & D. Stanković, 2023. A new perspective on the molecular dating of the brown trout complex with an extended phylogeographic information on the species in Serbia. Contributions to Zoology. https://doi.org/10.1163/18759866-bja10046.
Vera, M., P. Martinez & C. Bouza, 2018. Stocking impact, population structure and conservation of wild brown trout populations in inner Galicia (NW Spain), an unstable hydrologic region. Aquatic Conservation: Marine and Freshwater Ecosystems 28: 435–443.
Weiss, S., C. Schlötterer, H. Waidbacher & M. Jungwirth, 2001. Haplotype (mtDNA) diversity of brown trout Salmo trutta in tributaries of the Austrian Danube: massive introgression of Atlantic basin fish – by man or nature? Molecular Ecology 10: 1241–1248. https://doi.org/10.1046/j.1365-294X.2001.01261.x.
Weiss, S. & N. Ferrand (eds.), 2002. Phylogeography of Southern European Refugia: Evolutionary Perspectives on the Origins and Conservation of European Biodiversity. Springer, Dordrecht: 377 pp.
Weiss, S. & T. Schenekar, 2021. Kontrollbefischungen zum Regenbogenforellenvorkommen m Einzugsgebiet des Großen Baches – Nationalpark Kalkalpen. Final Report, University of Graz, Graz: 25 pp. [in German]
Weiss, S., L. Lecaudey & A. Wunder, 2017. Genetische Untersuchungen der Bachforelle in Fließgewässern des Nationalpark Kalkalpen. Final Report. University of Graz, Graz: 42 pp. [In German].
Whitely, A. R., K. Hastings, J. K. Wenburg, C. A. Frissel, J. C. Martin & F. W. Allendorf, 2018. Genetic variation and effective population size in isolated populations of coastal cutthroat trout. Conserveration Genetics 11: 1929–1943. https://doi.org/10.1007/s10592-010-0083-y.
Acknowledgements
We thank R. Haunschmid and F. Keil for providing the samples from the Kalkalpen National Park and for their valuable support to the project. We also thank the Tyrolean Fisheries Association, Austrian Federal Forests and M. Schletterer (TIWAG) for providing some of the samples. The Styrian samples come from the “Troutcheck” project, which was funded by the Province of Styria, Lower Austria and the Federal Ministry of Agriculture, Forestry, Environment and Water Management. Bavarian samples were collected with the logistic support and permits from the Lower Bavarian Fisheries Agency (S. Paintner), and U. Schliewen benefitted from financial support by a grant from the Stiftung für Artenschutz und Technik and the SNSB-Innovative program. Part of the Rhine samples come from the project “Erfassung und Dokumentation der genetischen Variabilität von Wildpopulationen der Bachforelle (Salmo trutta fario) aus verschiedenen Flussgebietseinheiten in Deutschland”, financed by the German Federal Ministry of Food and Agriculture (BMEL) through the Federal Office for Agriculture and Food (BLE), grant number BLE 12BE‐001. Finally, we would like to thank J. Grimm, A. Wunder and S. Schäffer (University of Graz) for lab assistance in library preparation. The authors acknowledge the financial support by the University of Graz.
Funding
Open access funding provided by University of Graz. This work was supported by a grant from the Doctoral Academy Graz, Ecology and Evolution in Changing Environments (EECE), University of Graz to G. Englmaier and the Austrian Center for Limnology (ACL, University of Graz, Austria), in the form of a PhD grant to L. Lecaudey. Field work and laboratory work were also supported by the Kalkalpen National Park through a joint grant with the Federal Ministry of Agriculture, Forestry, Environment and Water Management, and the European Union (LE14-20); and the Austrian Federal Forests (Österreichische Bundesforste) with a special thanks to A. Haas.
Author information
Authors and Affiliations
Contributions
Conceived and coordinated the study: GKE, LAL, and SJW. Laboratory work: LAL. Data analyses and methodology: GKE, LAL. Literature review and background information: GKE, LAL, UKS, ThS, TS, and SJW. Wrote the first draft of the paper: GKE. All authors contributed to the improvement of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling editor: Christian Sturmbauer
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Englmaier, G.K., Lecaudey, L.A., Schliewen, U.K. et al. Characterization of pure and admixed brown trout (Salmo trutta) populations of high conservation value in the upper Danubian contact zone using ddRADseq genotyping. Hydrobiologia 851, 2373–2388 (2024). https://doi.org/10.1007/s10750-023-05463-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05463-5