- 1Institute of Experimental Botany of the Czech Academy of Sciences, Prague, Czechia
- 2Department of Experimental Plant Biology, Faculty of Science, Charles University, Prague, Czechia
Plant secretory phospholipase A2 (sPLA2) is a family of lipolytic enzymes involved in the sn-2 hydrolysis of phospholipid carboxyester bonds, characterized by the presence of a conserved PA2c domain. PLA2 produces free fatty acids and lysophospholipids, which regulate several physiological functions, including lipid metabolism, plant growth and development, signal transduction, and response to various environmental stresses. In the present work, we have performed a comparative analysis of PA2c domain-containing genes across plants, focusing on gene distribution, phylogenetic analysis, tissue-specific expression, and homology modeling. Our data revealed the widespread occurrence of multiple sPLA2 in most land plants and documented single sPLA2 in multiple algal groups, indicating an ancestral origin of sPLA2. We described a novel PA2c-containing gene family present in all plant lineages and lacking secretory peptide, which we termed PLA2-like. Phylogenetic analysis revealed two independent clades in canonical sPLA2 genes referred to as α and β clades, whereas PLA2-like genes clustered independently as a third clade. Further, we have explored clade-specific gene expressions showing that while all three clades were expressed in vegetative and reproductive tissues, only sPLA2-β and PLA2-like members were expressed in the pollen and pollen tube. To get insight into the conservation of the gene regulatory network of sPLA2 and PLA2-like genes, we have analyzed the occurrence of various cis-acting promoter elements across the plant kingdom. The comparative 3D structure analysis revealed conserved and unique features within the PA2c domain for the three clades. Overall, this study will help to understand the evolutionary significance of the PA2c family and lay the foundation for future sPLA2 and PLA2-like characterization in plants.
1 Introduction
Phospholipases comprise an evolutionarily diverse group of lipolytic enzymes involved in membrane remodeling and hydrolysis of phospholipids into various bioactive lipid derivatives, including free fatty acids (FFA), phosphatidic acid (PA), diacylglycerol (DAG), and lysophospholipids (LPs) (Wang et al., 2012; Chen et al., 2013; Takáč et al., 2019). These lipid derivatives can regulate various physiological functions, including plant growth and development, cellular signaling, and stress management. Based on the site of phospholipid cleavage, phospholipases are classified into three main groups, phospholipase A (PLA), phospholipase C (PLC), and phospholipase D (PLD), each of them exhibiting further variations in the structure, regulation, and catalytic activity. PLA group, which is involved in the hydrolysis of carboxyester bonds from sn-1, sn-2, or both positions, can be further categorized according to the structural features, catalytic specificity, and calcium (Ca2+) requirement, as PLA1 (sn-1 carboxyester bond hydrolysis), secretory PLA2 (sn-2 carboxyester bond hydrolysis), and patatin-like PLA2 (hydrolyze both carboxyester bonds at sn-1 and sn-2 positions) (Chen et al., 2013; Takáč et al., 2019). PLA1 is calcium-independent, has molecular masses ranging from 45–50 kDa, consists of conserved GXSXG motif, and has a catalytic triad (Serine (S), Aspartic acid (D), and Histidine (H)) (Chen et al., 2013). The Arabidopsis genome encodes fourteen PLA1 gene members that regulate diverse functions such as plant growth and development, shoot gravitropism, production of jasmonic acid, senescence, and ultraviolet B (UV-B) defense signaling (Ishiguro et al., 2001; Kato et al., 2002; Lo et al., 2004; Hyun et al., 2008; Seo et al., 2008; Ellinger et al., 2010; Seo et al., 2011. Similarly, patatin-like PLA (pPLA) is a large enzyme with a patatin domain that serves as a prime active site and requires Ca2+ for catalytic activity (Chen et al., 2013). Arabidopsis encodes thirteen pPLA members involved in several physiological processes, including lipid metabolism, signal transduction, cell growth, and plant responses to biotic and abiotic stresses (La Camera et al., 2005; Yang et al., 2007; La Camera et al., 2009; Li et al., 2011; Yang et al., 2012; Li et al., 2013). Patatin-like PLA2s share a resemblance with the cytosolic animal iPLA2s (group VI) (Balsinde and Balboa, 2005). The other type of phospholipase A2, secretory PLA2 (sPLA2) is a small class of enzymes with low molecular masses ranging from 13–18 kDa, N-terminal secretory signal peptide, and PA2c domain (phospholipase A2; EC 3.1.1.4; SMART accession SM00085) that comprises a calcium-binding motif (YGKYCGxxxxGC) and a catalytic site DACCxxHDxC motif (Lee et al., 2005). The secreted sPLA2 variants are the most abundant PLA2s across nature and are classified into several subgroups I–III, V, IX–XIV. In plants, sPLA2 is evolutionarily grouped into XI IA (PLA2-β) and XI IB (PLA2-α) clades (Six and Dennis, 2000). It is involved in diverse physiological functions such as plant growth and development, cell elongation, gravitropism, regulation of auxin, and cellular signaling, and it was also implicated in the responses to biotic and abiotic stress (Takáč et al., 2019; Mariani and Fidelio, 2019).
The first successful plant sPLA2 enzyme purification and characterization attempt has been reported in developing elm seeds (Ulmus glabra) (Ståhl et al., 1998). Afterward, sPLA2 encoding cDNAs were isolated from shoots of Oryza sativa and flowers of carnation (Dianthus caryophyllus) (Kim et al., 1999; Ståhl et al., 1999). Later, various sPLA2 cDNA, genes, and proteins have been isolated and characterized from several plant species, including Arabidopsis thaliana, Solanum lycopersicum, Ricinus communis, Nicotiana tabacum, Triticum durum, Linum usitatissimum, and Citrus sinensis (Dhondt et al., 2000; Fujikawa et al., 2005; Liao and Burns, 2010; Fujikawa et al., 2012; Verlotta et al., 2013; Gupta and Dash, 2017). Arabidopsis genome encodes four sPLA2 isoforms (sPLA2-α, β, γ, and δ), that are categorized into two groups, alpha clade (sPLA2-α) and beta clade (β, γ, and δ). All three members of the beta clade (sPLA2-β, γ, and δ) have been shown to play essential roles in pollen development and pollen tube growth (Kim et al., 2011). Arabidopsis sPLA2-α is required for the trafficking of PIN-FORMED auxin efflux transporters to the plasma membrane and negatively regulates the plant’s defense response by repressing the AtMYB30 transcription activity during pathogen infection (Lee et al., 2010; Froidure et al., 2010). AtsPLA2-β produces second messengers to enhance light-induced stomatal opening and contributes to cell elongation and shoot gravitropism through the auxin signaling pathway (Lee et al., 2003; Seo et al., 2008).
Despite considerable work on the plant PLA2, there is little information on comparative structural studies of plants XI IA (PLA2-β) and XI IB (PLA2-α) members and no data are available on deep evolutionary comparisons within the plant kingdom (Mansfeld, 2009). Several eukaryotic sPLA2 crystal structures have been elucidated, whereas in plants, only the rice sPLA2 crystal structure has been solved (Guy et al., 2009). It demonstrated that six disulfide bonds stabilize the rice sPLA2 structure, where the N-terminus contains the conserved Ca2+-binding loop, which starts with a short 310-helix and two short antiparallel β-strands (Guy et al., 2009). Moreover, the C-terminus is folded into three antiparallel α-helices and contains the conserved catalytic histidine and aspartate (HD) residues (Guy et al., 2009). In the present study, we have analyzed the global distribution of sPLA2 members across the plant kingdom. We have identified deep-branching, previously uncharacterized PA2c domain-containing subfamily, termed PLA2-like, in plants. We analyzed the evolutionary relationship among sPLA2 and PLA2-Like members, which separated sPLA2-α, β and PLA2-like into three distinct clades. To get more insight into the expression of the three clades, we have compiled tissue-specific expression data for several angiosperm species, which demonstrated the PLA2-β members are mainly expressed in the male reproductive tissues. Promoter analysis predicted the presence of tissue, hormone, light, and stress-responsive cis-acting motifs. To uncover conserved structural features in the PA2c fold, we have performed comparative homology modeling of Amborella trichopoda, Arabidopsis thaliana, and Nicotiana tabacum TN90 PLA2-α, β, and PLA2-like members. Collectively, this study sheds new light on the sequence and structural evolution of the plant sPLA2 family.
2 Materials and methods
2.1 Plant sPLA2 homologs identification and annotations
A well-annotated Arabidopsis, rice sPLA2-α and β protein sequences were retrieved from Phytozome v13 (https://phytozome.jgi.doe.gov/pz/portal.html) and compiled as initial query sequences. These AtsPLA2 query sequences were used to perform BlastP search against thirty-four plant genomes, including Chlorophyta, Bryophyta, Pteridophyta, Gymnosperm, and Angiosperm species in Phytozome v 13 (https://phytozome.jgi.doe.gov/pz/portal.html), ONEKP database (One Thousand Plant Transcriptomes Initiative, 2019) and NCBI database. Moreover, sPLA2 sequences of several Nicotiana species (N. tabacum TN90, N. sylvestris, and N. tomentosiformis) were searched in the SolGenomics database (Mueller et al., 2005). In subsequent rounds of blast searches, PLA2 sequences from bryophytes, and charophyte and chlorophyte algae were also used as queries. Full-length protein and nucleotide sequences were retrieved and manually checked for the presence of an N-terminal secretory peptide, highly conserved PA2c domain in the ScanProsite (https://prosite.expasy.org/scanprosite/), and Interproscan 5 (https://www.ebi.ac.uk/interpro). Further, partial and truncated sequences that suggested incomplete gene predictions were curated using the SoftBerry FGENESH+ gene prediction algorithm (https://www.softberry.com); short and dubious sequences were removed from the dataset. All sPLA2 and PLA2-like sequences were compiled in the table with their genomic information, including gene id, protein length, and chromosomal location (Supplementary Table 1).
2.2 Sequence alignment and phylogenetic analysis
Protein sequences were aligned using the MAFFT E-INS-I algorithm (Katoh and Standley, 2013) in Jalview (Waterhouse et al., 2009). All sequences were manually checked for gaps and non-conserved regions, which were eliminated from the alignment and exported in FASTA file format. The resulting alignment was used to build a sequence logo of the Ca2+ binding motif and catalytic motif (https://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004). The evolutionary relationship between plant sPLA2 and PLA2-like was inferred using the IQ-TREE algorithm with Maximum likelihood (ML) supported by the ultrafast bootstrap method (1000 replicates). Model finder was performed and the Whelan and Goldman model with Invariable and gamma (WAG+I+G4) was selected as a best model based on the Bayesian information criterion (BIC) score (Minh et al., 2020). The Interactive Tree Of Life (iTOL v5) (https://itol.embl.de/) online tool was used to display and annotate the sPLA2 phylogenetic tree. Likewise, species trees were constructed in NCBI taxonomy and edited in the iTOL server.
2.3 Plant sPLA2 and PLA2-like genes cell and tissue-specific expression analysis
Tissue-specific expression of the sPLA2 and PLA2-like members have been searched in various vegetative (leaf, stem, root, and seeds) and reproductive (flower, anther, pollen, pollen tube, carpels, pistil, ovary, ovule, and egg cells) tissues of Arabidopsis, Amborella, tomato, grape, rice, and maize using the CoNekT database (https://conekt.sbs.ntu.edu.sg/) (Proost and Mutwil, 2018). Gene expression was represented in transcripts per kilobase million (TPM)-based normalization because it can be used for both gene count comparisons within a sample or between samples of the same sample group (Abrams et al., 2019). The expression values were analyzed in the CIMminer one matrix server (discover.nci.nih.gov/cimminer).
Total RNA was isolated from tobacco leaves, roots, buds, flowers, imbibed pollen, germinating pollen grains and growing pollen tubes using Qiagen RNAeasy Kit, and Turbo DNA-free Kit (Applied Biosystems, Waltham, MA, USA) was used for DNA removal. cDNA synthesis was carried out using Transcriptor High Fidelity cDNA Synthesis Kit (Roche, Penzberg, Germany) with anchored-oligo (DT)18 primer according to manufacturer’s instructions. Semi-quantitative RT-PCR was performed with NtPLA2 gene-specific oligonucleotides 1-6 (Supplementary Table 2) designed to span an intron in the corresponding genomic DNA sequence. Actin7 (Bosch et al., 2005) was used as load control. Amplification conditions were 94°C for 30 sec, 55°C for 30 sec, 68°C for 30 sec and final extension 68°C for 10 min for 28 or 34 cycles.
2.4 Cis-acting elements prediction in sPLA2 and PLA2-like promoters
The promoter regions of sPLA2 and PLA2-like genes (~ 2kbp upstream of the start codon) were retrieved from the Phytozome v13 database. Promoter sequences were then analyzed via the PlantCARE server with default parameters (http://bioinformatics.psb.ugent.be/webtools/plantcare/html). The obtained cis-acting elements data were processed using the CIMminer server.
2.5 Prediction of homology models of the sPLA2 and PLA2-like proteins
Homology models of N-terminal truncated peptides signals of Amborella trichopoda (ATR0564G112-α; 129 aa, ATR0789G151-β; 118 aa), Arabidopsis thaliana (AT2G06925-α; 128 aa, AT2G19690-β; 119 aa), and Nicotiana tabacum TN90 (Gene_60450(SS4740)-α; 127 aa, Gene_37244 (SS1768)-β; 117 aa) were obtained through AlphaFold server and also predicted using Robetta server (https://robetta.bakerlab.org/), and RaptorX server (http://raptorx.uchicago.edu/). Moreover, homology models of C-terminal regions (containing PA2c domain) of PLA2-like sequences were predicted for Amborella trichopoda (AmTr _scaffold00063; 168 aa), Arabidopsis thaliana (AT4G29070- 145 aa), and Nicotiana tobacumTN90 (mRNA_158062; 149 aa). For Robetta and RaptorX, five models of each protein were generated, and the model qualities were compared with the AlphaFold predictions using the SWISS-refinement server (https://swissmodel.expasy.org/assess). MolProbity Score, clash score, Ramachandran scores, and QMEAN were considered to select the best model among all generated predictions. Further, electrostatic potentials for the best models were calculated using APBS web-server (Adaptive Poisson-Boltzmann Solver, https://www.poissonboltzmann.org/) and plotted on the predicted protein structures (Unni et al., 2011). The conserved residues and non-conserved residues were analyzed using the Consurf web-server (https://consurf.tau.ac.il/) for the best models. All the generated models were visualized and edited using the CHIMERA (https://www.cgl.ucsf.edu/chimera/).
3 Results
3.1 Phylogenomic analysis of plant PA2c domain-containing genes revealed an ancient origin of sPLA2 and uncovered a widespread presence of the previously uncharacterized PLA2-like family
To get a deeper insight into the evolution of the plant sPLA2 family, we have selected thirty-four plant genomes representing a diverse and balanced sample of Viridiplantae, including algae, bryophytes, lycophytes, pteridophytes, gymnosperms, and angiosperms. We searched within this genome sample for the presence of the sPLA2 homologs, using known dicot and monocot sPLA2 as input and employing various homology-based searches like Blast and HMMER (Figure 1A). We ultimately identified 113 non-redundant canonical plant sPLA2 genes based on the following criteria: the presence of N-terminal signal peptide and PA2c domain with Ca2+ binding motif and catalytic HD dyad site. Surprisingly, we also uncovered 37 additional genes coding for proteins that lack the N-terminal signal peptide but contain a well-conserved PA2c domain, including the calcium-binding motif and the catalytic dyad (Supplementary Figure 1, Supplementary Tables 1, 3). The data showed a ubiquitous distribution of both sPLA2 and PLA2-like genes in plants, from unicellular algae to multicellular flowering plants (Figure 1A). Remarkably, the two groups exhibit a clear difference in evolutionary dynamics. PLA2-like orthologs were found as single-copy genes in most diploid species and never exceeded two copies per genome. Two PLA2-like genes were found in either polyploid species (N. tabacum) or species that underwent relatively recent whole genome multiplications (B. rapa, G. max, P. patens). This suggests that PLA2-like gene number is under the purifying selection to remain singleton. On the other hand, sPLA2 genes display much wider genomic plasticity, showing gene numbers ranging from one (chlorophyte and streptophyte algae) to twelve (gymnosperm T. plicata). This higher evolutionary dynamics of sPLA2 is also apparent for individual plant clades like ferns (two to nine genes) and gymnosperms (four to twelve genes). Within angiosperms, Basal lineages (represented by A. trichopoda) seem to contain two sPLA2 genes, while distinct dynamics can be found between dicots (two to six genes) and monocots (four to five genes). Despite the general increase in sPLA2 gene complexity during plant evolution, reductive events can also be observed. In bryophytes, moss P. patens and liverwort M. polymorpha have two sPLA2 isoforms, whereas only one sPLA2 member was observed in hornwort A. angustus. Similarly, just one sPLA2 was retained in the lycophyte S. moellendorffii (Figure 1A).
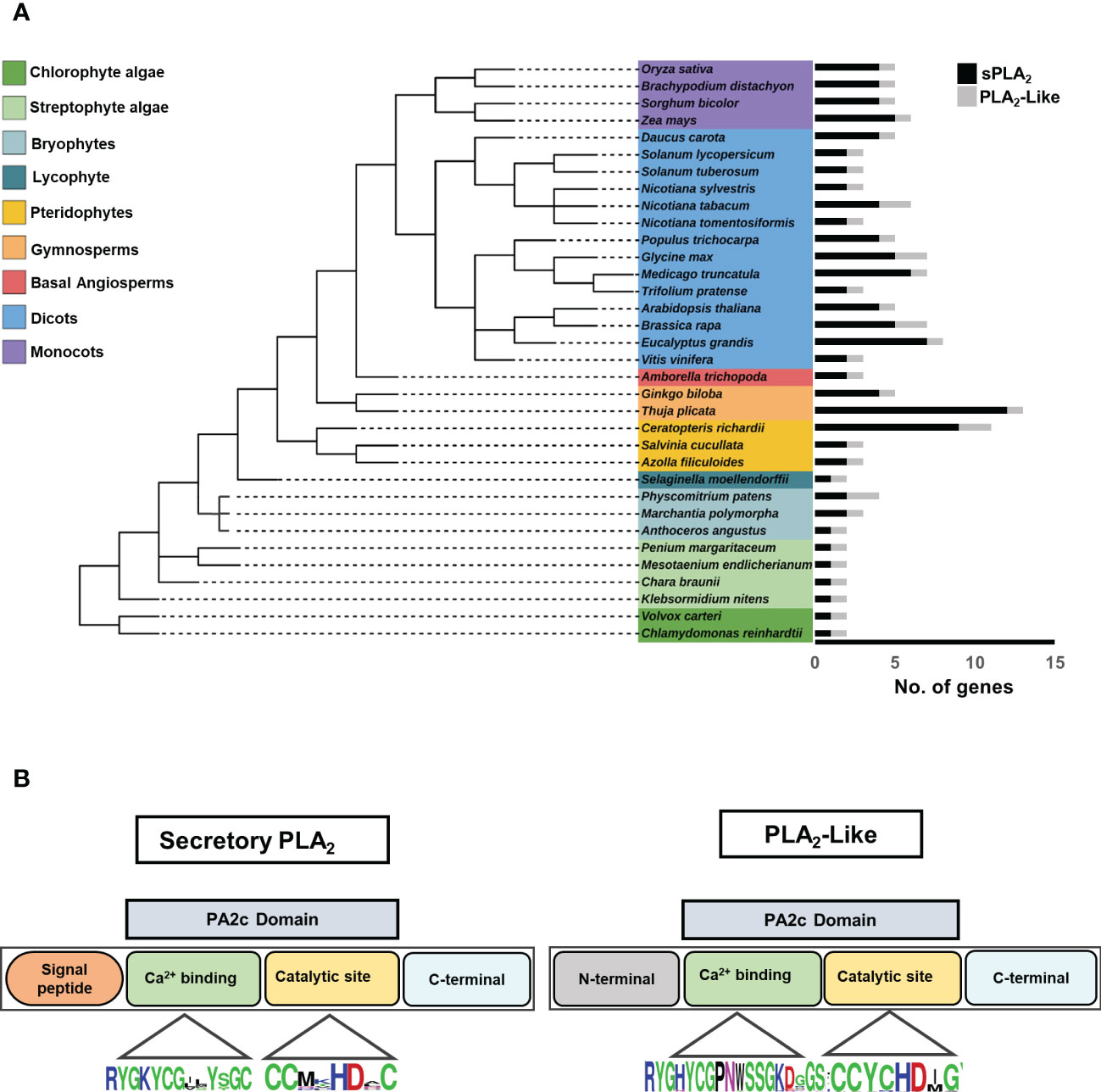
Figure 1 (A) Distribution of secretory PLA2 and PLA2-like genes in the different plant taxonomic groups, including Algae, Bryophytes, Pteridophytes, Gymnosperms, and Angiosperms. The species tree was elucidated using the NCBI taxonomy database and edited in the Interactive Tree of Life (iTOL) web tool. Each histogram corresponds to the number of sPLA2 and PLA2-like in each species. Black, histogram-sPLA2; Gray, histogram-PLA2-like genes. (B) Schematic representation of PLA2 protein with N and C terminal, conserved PA2c domain, Ca2+ binding motif, and HD catalytic dyad.
To get more insight into the sequence characteristics of sPLA2 versus PLA2-like subfamilies, we performed multiple sequence alignments of the two groups and analyzed their overall protein sizes, motif occurrences, and the distribution of conserved residues. All sPLA2 sequences have N-terminal signal peptides and range between 90-191 residues, while PLA2-like members range between about 143-320 residues, have extended N- and C-terminal regions, and lack any recognizable signal peptide sequence.
The sequence analysis revealed other notable differences among sPLA2 and PLA2-like members at amino acid levels. Significantly, all sPLA2 members possess twelve cysteine residues forming six disulphide bridges and providing extra structural stability (Mariani and Fidelio, 2019). In contrast, PLA2-like members have only six C residues, possibly forming up to three disulphide bridges (Supplementary Table 3). Moreover, although the highly conserved Ca2+ binding loop is present in all members within the PA2c domain, PLA2-like has a subtle variation in Ca2+ binding motif with the insertion of an extra residue (YGHYCGxxxxxGK vs YGKYCGxxxxGC) (Figure 1B). Another cysteine loss occurred near the catalytic site where sPLA2-α and β show invariable DxCCxxHDxC motif, whereas PLA2-like has DxCCxxHDxG.
Collectively, our data show a widespread and dynamic occurrence of canonical sPLA2 genes and document a highly conserved, evolutionary-constrained subfamily of non-characterized PLA2-like genes.
3.2 Phylogenetic analysis revealed that sPLA2 and PLA2-like sequences clustered into three deep-branching clades
To shed light on the evolutionary relationships of sPLA2 and PLA2-like families, a curated dataset of 113 sPLA2 and 38 PLA2-like genes was compiled from a diverse set of plants with sequenced genome drafts (Figure 2). It included evolutionary important chlorophytic and streptophytic algal species such as Chlamydomonas reinhardtii, Volvox carteri, Penium margaritaceum, Klebsormidium nitens, Mesotaenium endlicherianum, and Chara braunii. Moreover, we have included several bryophytes (moss Physcomitrium patens, liverwort Marchantia polymorpha, and hornwort Anthoceros angustus), lycophyte (Selaginella moellendorffii), pteridophytes (Azolla filiculoides, Salvinia cucullata, and Ceratopteris richardii), gymnosperms (Thuja plicata and Ginkgo biloba), Basal angiosperm (Amborella trichopoda), and multiple dicots, and monocots species (Figure 2). The sequences were aligned using the MAFFT E-INS-I algorithm, unalignable regions were removed, and a maximum likelihood (ML) tree was constructed using the IQ-tree software with ultra-fast bootstrap support (Figure 2).
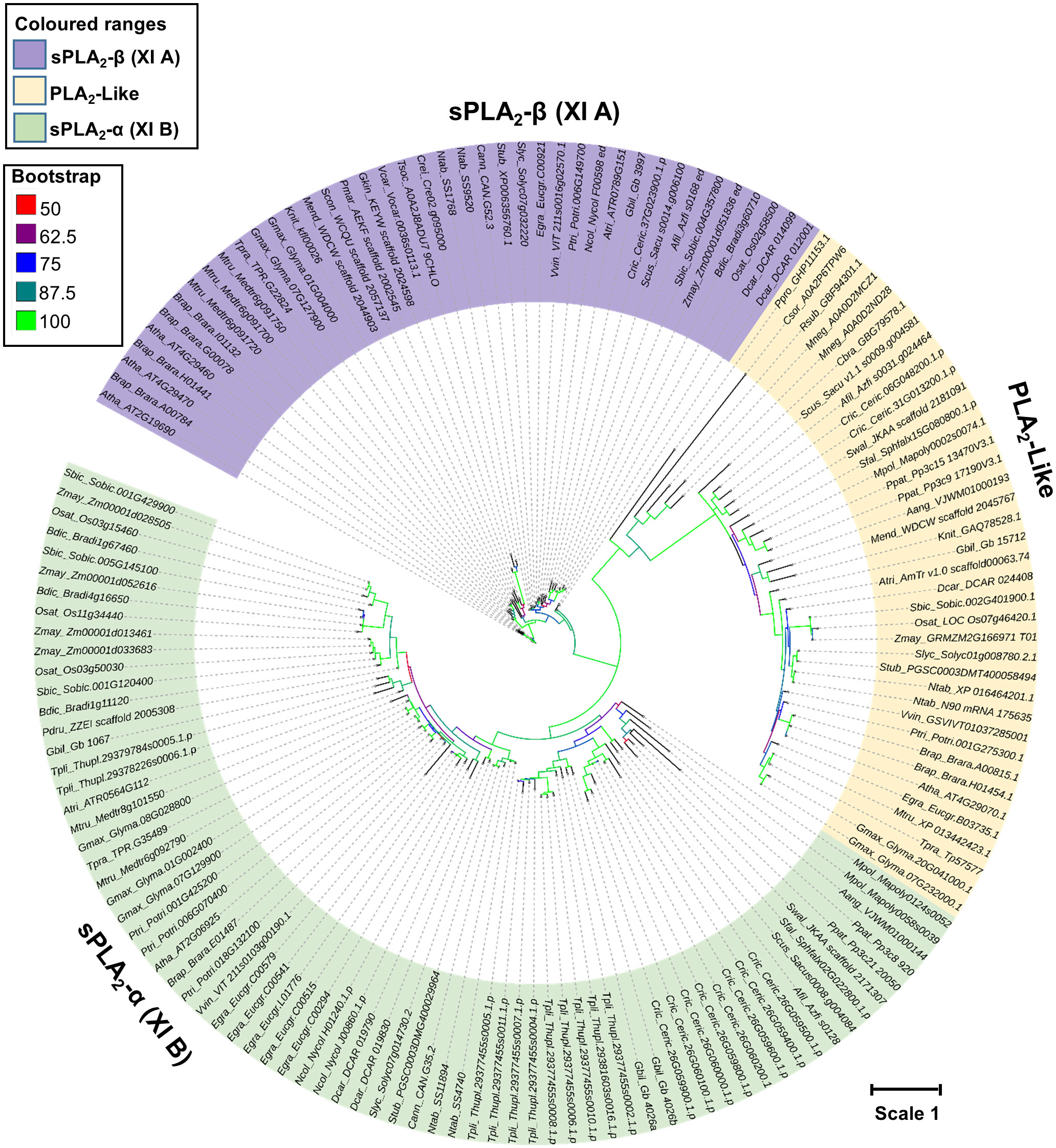
Figure 2 Phylogenetic relationships between sPLA2 and PLA2 -like members were constructed using the Maximum likelihood (ML) with the ultrafast bootstrap method. The evolution Whelan and Goldman model with Invariable and gamma (WAG+I+G4) was selected based on the Bayesian information criterion (BIC) score. Color nodes correspond to the bootstrap support. The scale bar indicates the rates of substitutions/site. Atha, Arabidopsis thaliana; Afil, Azolla filiculoides; Aang, Anthocerus angustus; Atri, Amborella trichopoda; Brap, Brassica rapa; Bdic, Brachypodium distachyon; Cann, Capsicum annuum; Crei, Chlamydomonas reinhardtii; Csor, Chlorella sorokiniana; Cric, Ceratopteris_richardii; Dcar, Daucus carota; Egra, Eucalyptus grandis; Gbil, Ginkgo biloba; Gmax, Glycine max; Gkin, Gonatozygon_kinahanii; Knit, Klebsormidium nitens; Mneg, Monoraphidium neglectum; Mpol, Marchantia polymorpha; Mtru, Medicago truncatula; Mend, Mesotaenium endlicherianum; Ntab, Nicotiana tabacum; Ncol, Nymphaea colorata; Osat, Oryza sativa; Pdru, Phylloglossum drummondii; Ptri, Populus trichocarpa; Pmar, Penium margaritaceum; Ppat, Physcomitrium patens; Ppro; Pycnococcus provasolii; Slyc, Solanum lycopersicum; Stub, Solanum tuberosum; Sbic, Sorghum bicolor; Tpli, Thuja plicata; Rsub, Raphidocelis subcapitata; Scon, Staurodesmus_convergens; Swal, Selaginella_wallacei; Sfal, Sphagnumfallax; Scus, Salvinia cucullata; Tsoc, Tetrabaena socialis; Tpra, Trifolium pratense; Vvin, Vitis vinifera; Vcar, Volvox carteri; Zmay, Zea mays.
Despite a limited evolutionary signal (due to relatively short sequence lengths), the resulting tree showed clearly that plant PA2c domain-containing genes separated into three well-supported clades. All sPLA2 sequences were clustered into two groups, referred to as sPLA2-α clade and β clade and corresponding to XI-A (sPLA2-β) and XI-B (sPLA2-α) classification of sPLA2 genes (Mansfeld, 2009; Mariani and Fidelio, 2019). Significantly, PLA2-like members formed a third independent clade, possibly separating sPLA2-α and sPLA2-β (Figure 2). It should be noted that the two canonical clades seem to possess several specific enzymological characteristics, which were best explored in Arabidopsis and elm. Most importantly, sPLA2-α members are show optimal activation by millimolar calcium, while sPLA2-β show maximal activity already at micromolar calcium levels. Compared to sPLA2 members, newly identified PLA2-like members are large in length (>240 aa), high molecular weight, disordered N-terminal region (~100 aa) without signal peptide, and residual variation in Ca2+ binding loop. Moreover, the presence of clear sPLA2-α and sPLA2-β orthologs in gymnosperms and ferns clearly shows that the diversification and stable retention of an α- and β-clade occurred relatively early in land plant evolution. Interestingly, the contrasting sPLA2 distribution in algae and bryophytes (all chlorophyte and streptophyte algae retain 1 sPLA2 isoform clustering in the β-clade, while mosses, liverworts and hornworts possess only α-clade members), suggests that the two clades may have emerged very early in plant sPLA2 evolution, and the single clades were lost in distinct lineages. On the whole, the evolution of the β-clade is clearly under the evolutionary constraint, both in gene number (except for Brassicaceae) and mutation rate (Figure 2). On the other hand, rapidly evolving α-clade is highly expanded in several lineages, including pteridophytes (C. richardii), gymnosperms (T. plicata and G. biloba), and angiosperms (E. grandis, P. trichocarpa, G. max, and M. truncatula).
As mentioned above, PLA2-like clade, which consists of primarily single-copy genes, branched-off from sPLA2 at the earliest stage of green plant evolution, although its presence in green algae was detected universally (Supplementary Table 1). Generally, PLA2-like phylogeny roughly follows the species evolution with a mutational rate similar to sPLA2-α, suggesting a gradual evolution constrained at the gene number level (Figure 2).
3.3 Global expression analysis shows the overlapping expression of sPLA2 and PLA2-like genes in the sporophyte and suggests a dominant presence of sPLA2-β clade members in the male gametophyte
To get a comprehensive evolutionary insight into sPLA2 expression, publicly available tissue-specific RNA-Seq data of representative members of eudicots (A. thaliana, S. lycopersicum, and V. vinifera), monocots (O. sativa and Z. mays), and Basal angiosperms (A. trichopoda) have been extracted and arranged clade-specific on the phylogenetic tree (Figure 3A). In total, we analyzed the expression of eleven α-clade members, eight β-clade members, and six PLA2-like members in four vegetative tissues (leaves, stem, root, and seeds) and ten reproductive tissues (flower, anther, pollen, pollen tube, carpels, pistil, ovary, ovules, and egg cells). The data strongly suggest the existence of conserved clade-specific expression patterns across angiosperms. Members of the sPLA2-α clade in all six analyzed species exhibited significant expression in the vegetative tissues, including leaf, stem, root, and seeds. Notably, while most dicot, monocot and Amborella sPLA2-α members are expressed in the various maternal reproductive tissues, such as flowers, anther, carpels, ovules, and egg cells, they are all conspicuously absent from pollen and pollen tube.
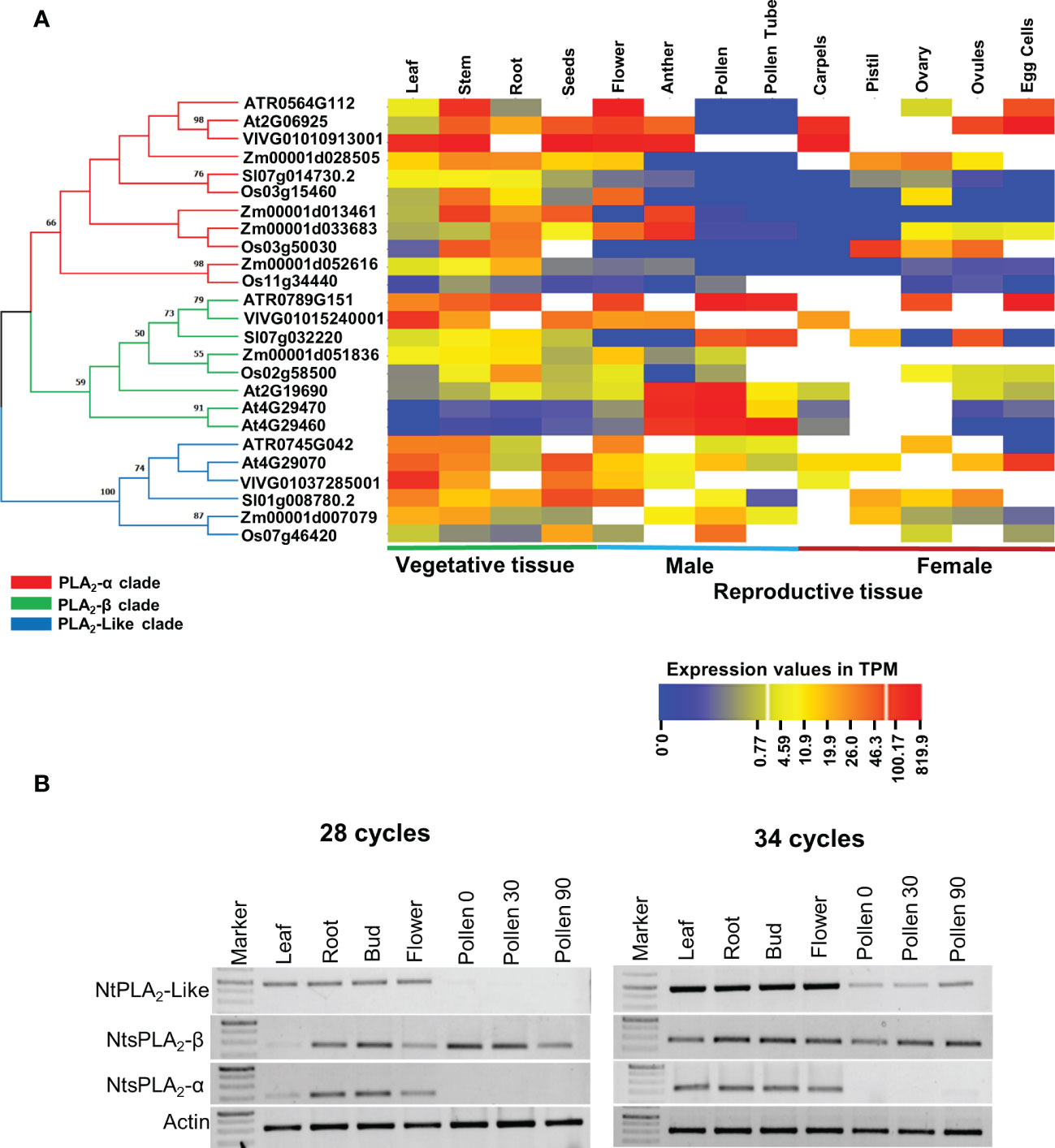
Figure 3 (A) Vegetative and reproductive tissue-specific gene expression data of sPLA2 and PLA2-like members were collected for the selected plant species and arranged on a phylogenetic tree. All collected data were exploited to generate the heatmap using the CIMminer web tool. The expression data was represented as a transcript per million (TPM). The phylogenetic relationships between sPLA2- α, β, and PLA2-like members were determined using an ML tree with 100 bootstrap replications in the MEGA X software with the default model. (B) Semi-quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis showing the expression of tobacco sPLA2 and PLA2-like members in different tissues and during pollen germination (Pollen30) and pollen tube elongation (Pollen90).
On the other hand, sPLA2-β members across angiosperms showed significant (often the strongest) expression in male gametophytic tissues such as pollen, and pollen tube (and also anther), in addition to variable sporophytic expression. This is well illustrated on the three β-clade and one α-clade Arabidopsis members, where our global expression data analysis is corroborated by earlier RT-PCR and promoter studies (Bahn et al., 2003; Wang et al., 2008). Thus, AtsPLA2-α and β show similar expression in all tissues except male gametophyte, where AtsPLA2-β is significantly expressed. Conversely, AtsPLA2-γ (and to a lesser extent also AtsPLA2-δ) displayed significant expression predominantly in the male gametophyte tissues, including anther, pollen and pollen tube, suggesting that these isoforms might play an essential role in plant reproduction (Bahn et al., 2003; Lee et al., 2005; Wang et al., 2008). The general expression profile of angiosperm PLA2-like then suggests a ubiquitous expression with the highest values typically occurring in the sporophyte and species-diversified expression in the male and female gametophytes (Figure 3A).
To experimentally corroborate these in silico studies, we performed a semiquantitative RT-PCR analysis of tobacco PA2c homologs (please note that the sequences of tobacco sPLA2 and PLA2-like homoeologous gene pairs are nearly identical (> 98% nucleotide identity) in tobacco amphidiploid genome and were considered as single cDNAs in the RT-PCR analysis). Analysis of leaf, root, bud, flower, dry pollen, germinating pollen and growing pollen tubes showed that tobacco PA2c genes show expression patterns strongly supporting the RNAseq data on other species, particularly the absence of AtsPLA2-α in pollen and pollen tubes, where PLA2-β is strongly present (Figure 3B). Similarly, tobacco PLA2-like was expressed predominantly in the sporophyte but could be clearly detectable also in the male gametophyte.
Taken together, our comprehensive examination of available transcriptomic data of six angiosperm species and RT-PCR analysis of sPLA2 and PLA2-like genes in tobacco showed evolutionarily conserved expression patterns for the three clades, highlighting their overlapping expression in most sporophytic tissues, and suggesting a significant role for the sPLA2-β clade in pollen.
3.4 Presence of light, stress, and hormone-responsive elements in sPLA2 and PLA2-like promoters across genomes
Having established conserved expression patterns for the three clades of the angiosperm PLA2 family, we next sought to understand their transcription regulatory networks. Therefore, we predicted cis-elements for the promoter sequences of sPLA2 and PLA2-like genes from Arabidopsis, wine, rice, and Amborella (Figure 4). Various categories of cis-elements were found, including common elements TATA and CAAT box and tissue-specific, light, stress, and hormone-responsive elements.
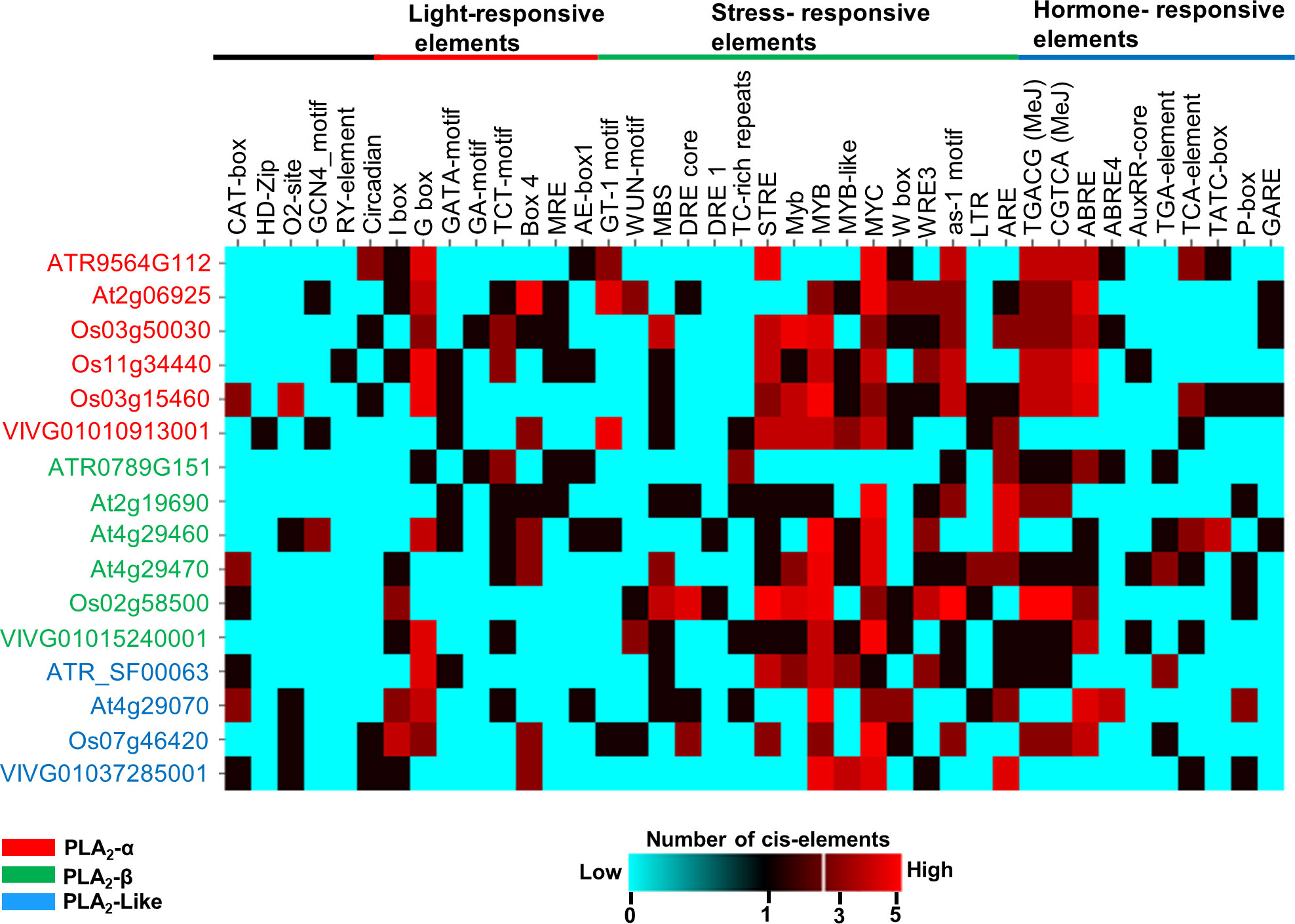
Figure 4 In-silico prediction of cis-acting elements in the promoter region (5’ upstream region about 2kbp) using PlantCARE database. The number of elements in the promoter regions was depicted by a heatmap using the CIMminer web tool. The color code represents the number of cis-elements in the promoter regions; Cyan- 0 (absent), Red- 5 copies of the element.
While no obvious pattern of evolutionarily conserved subset of cis-acting motifs could be attributed to individual sPLA2 or PLA2-like clades, the analysis strongly suggested that specific stress- and hormone-responsive elements are the most significantly overrepresented category in angiosperm sPLA2 and PLA2-like genes. These include abiotic stress-responsive elements, like dehydration-responsive element MBS (TAACTG), Myb (CAACTG), MYB (CAACAG), MYB recognition sites (CAACAG), MYB-like sequence (TAACCA) and MYC (CATTTG).
In addition to the abiotic stress-related elements, motifs implicated in biotic interactions, such as wound-responsive elements W-box (TTGACC), WRE3 (CCACCT), and as-1, were detected in most promoter regions for distinct species. Among those, the WRE3 motif is the most abundant and reported in almost all sPLA2-α and β gene members except Amborella and grape genes. W-boxes were found to interact with transcription factors belonging to the WRKY family, regulate defence-related genes, and play a vital role in biotic and biotic stress, senescence and seed dormancy.
Several hormone-responsive elements were documented in the promoter regions, among which abscisic acid-responsive element ABRE (TACGTG) and methyl jasmonate-responsive cis-elements (CGTCA and TGACG) are the most abundant and conserved, particularly within the sPLA2-α clade. On the other hand, ethylene-responsive element ERE (ATTTCAAA), gibberellic acid-responsive elements (GARE: AAACAGA, PA box, and TATC box), auxin responsiveness core element (auxRR: GGTCCAT, TGA elements), show much lower abundance and are scattered among species (Figure 4).
Lastly, many light-responsive cis-elements have been documented in the sPLA2 and PLA2-like promoter regions, including I-box, G box, GATA-motif, GA motif, TCT motif, Box 4, and MRE (MYB binding light responsive elements). The most abundant and conserved motif is G-box (CACGTG), which is involved in the light, abscisic acid, methyl-jasmonate, ethylene and anaerobiosis responses (Sibéril et al., 2001).
Collectively, the abundantly predicted cis-acting elements corroborate earlier functions experimentally attributed to selected sPLA2 members and suggest a conserved transcriptional control. Indeed, sPLA2 transcription was activated in response to blue light (Seo et al., 2008), auxin (Scherer, 2002; Lee et al., 2010), abiotic stresses (Chapman, 1998), wound stress and pathogen elicitors (Creelmen and Mullet, 1997; Lee et al., 1997; Laxalt and Munnik, 2002; Ellinger et al., 2010).
3.5 Comparative analysis of sPLA2 and PLA2-like structural models
Despite a wealth of knowledge about PLA2 physiological function, there is limited structural data on the plant sPLA2-α, β, and PLA2-like members, thus impeding our understanding of sPLA2 substrate preferences, interfacial recognition surface (IRS), and catalytic sites. To get better insight into the structural features of sPLA2 and PLA2-like proteins, we thoroughly analyzed structural models predicted for representative members of sPLA2-α, β, and PLA2-like from Arabidopsis, tobacco, Amborella (Figure 5; Supplementary Figure 2, 3). Since the current databases of automatically-generated structural models include the signal peptide, we generated de novo models of the processed sPLA2 forms. Moreover, since the N-terminal extensions in PLA2-like sequences show the characteristics of an intrinsically disordered region, we analyzed only the C-terminal portions. In a pilot analysis, we generated the models using three top-ranked algorithms that are not constrained by existing experimental structures (AlphaFold2_mmseq2 via the ColabFold infrastructure, RoseTTAFold, and RaptorX). We assessed them for their folding accuracy based on the Molprobity score, Ramachandran score and Q means plus Z-score criteria (Wang et al., 2016; Waterhouse et al., 2018; Jumper et al., 2021; Baek et al., 2021). Almost universally, AlphaFold-generated models displayed the best criteria and were therefore selected for further analyses (Supplementary Table 4).
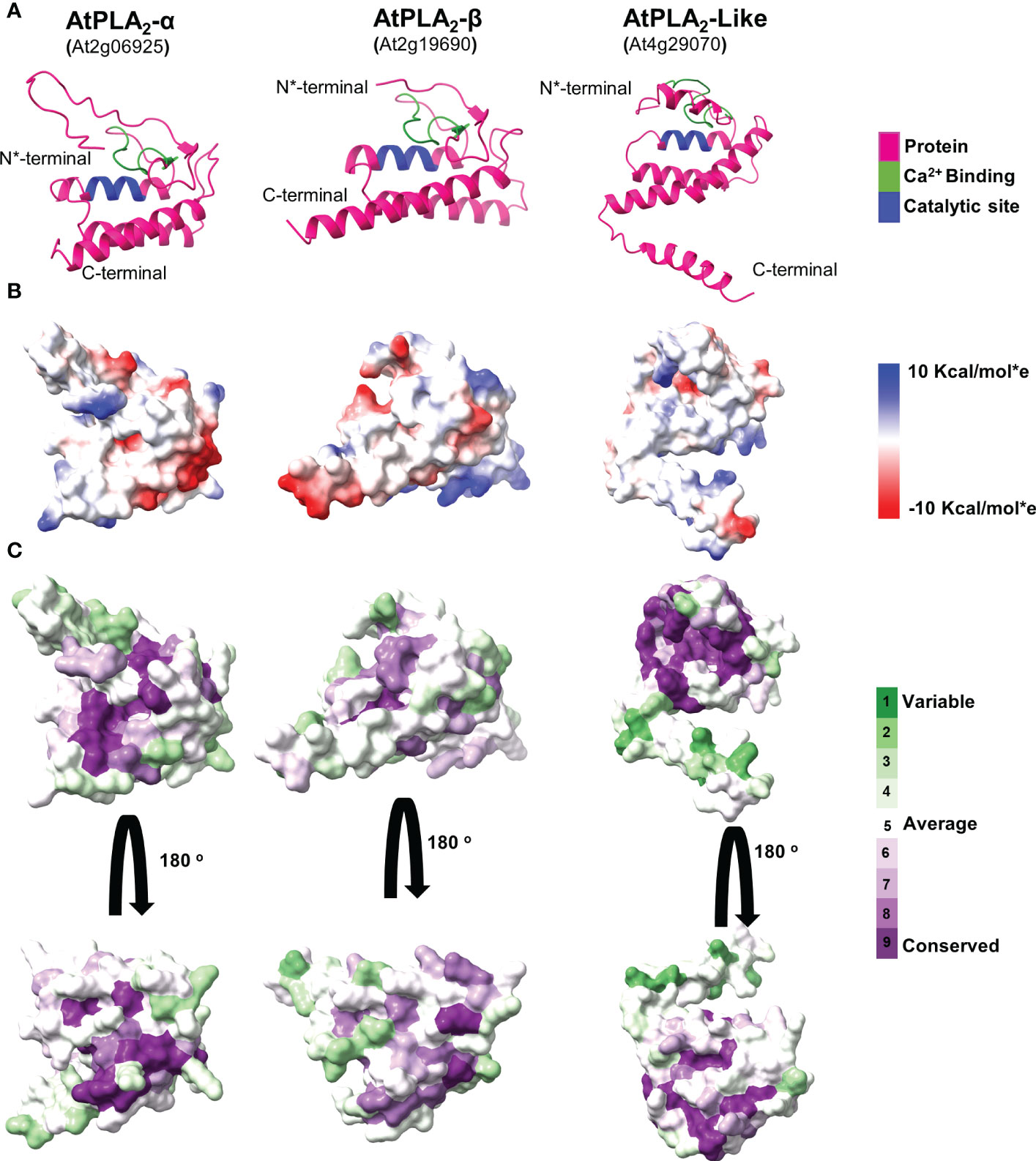
Figure 5 A comparative analysis of predicted Arabidopsis PLA2 structural models. (A) Cartoon representation of sPLA2-α, PLA2-β, and PLA2-like protein models. (B) A comparative analysis of the Coulombic electrostatic potential distribution among PLA2 members, calculated with the ChimeraX software. (C) Analysis of conserved residues on the sPLA2 and PLA2 surfaces, calculated using the Consurf server.
Comparative analysis of the final validated models revealed that all display a rather tightly-packed globular structure corresponding to the general sPLA2 fold (Figure 5A; Supplementary Figure 2, 3). Two main structural regions are present across the analyzed species in all sPLA2 and PLA2-like models. The N-terminal part contains mainly a loop region, including a conserved Ca2+-binding loop, and is preceded by two antiparallel beta strands. The C-terminal segment is then represented by three antiparallel α-helices, of which the two first are also present in other secreted PLA2s and contain the conserved catalytic histidine and calcium-coordinating aspartate residues (Guy et al., 2009). Similar structural features were documented in PLA2-like sequences, including the N-terminal loop region with Ca2+ binding motif and three antiparallel helices at the C-terminal region with the extended region. Notably, in canonical sPLA2s, the calcium-binding loop and the N-terminal and C-terminal parts are held together by six disulfide cysteine bridges that stabilize the overall structure. However, not all cysteines are conserved in PLA2-like sequences, and the structural models suggest that only two cysteine bridges may be retained in PLA2-like structures, giving them higher conformational flexibility.
Since sPLA2 is an interfacial enzyme interacting with the phospholipid bilayer, we next sought to see the electrostatic potential distribution in all analyzed models that could indicate distinct membrane-binding patterns for separate clades. Our data show that sPLA2-β members in all analyzed species contain positively-charged pockets, which may be involved in anionic lipid binding. On the other hand, all PLA2-like models lack this feature and produce mostly neutral surfaces. The highest diversity was found within sPLA2-α clade, where significant charge differences can be found between Arabidopsis, Nicotiana and Amborella members (Figure 5B; Supplementary Figure 2, 3). The low evolutionary conservation of the charge distribution was also corroborated by the visualization of evolutionarily conserved surface residues, suggesting that among sPLA2-α, β, and PLA2-like members, there is rather low conservation beyond the Ca2+ binding motif and the catalytic region (Figure 5C).
4 Discussion
4.1 Ubiquitous distribution and ancient divergence record of sPLA2 in plants
Despite the critical importance of PLA2 documented in higher plants and the availability of detailed molecular information for phospholipases from non-plant organisms, evolution-based knowledge of phospholipase A2 from plants is still meagre. To fill this gap, we have performed a comparative phylogenomic, sequence and structural analysis of the plant’s secretory PLA2 family, along with previously uncharacterized PLA2-like members. This is the first systematic analysis of the plant family of PA2c domain-containing proteins, covering chlorophyte and streptophyte algal species, bryophytes, lycophyte, pteridophytes, gymnosperms, Basal angiosperm, dicots and monocots. However, the detailed evolutionary history and clade separation of the plant PA2c domain superfamily is murky. Our data demonstrated that one genuine sPLA2 can already be documented in multiple algae species (chlorophytes, charophytes and zygnematophytes), and all canonical algal sPLA2 fall into the XI-A (sPLA2-β) clade. On the other hand, all canonical sPLA2 genes from three main bryophyte groups (hornworts, liverworts, and mosses) belong to the XIB (sPLA2-α) clade, and the simultaneous presence of both clades can be first documented in ferns. Three evolutionary scenarios can explain this diverse distribution: (i) sPLA2-β is the ancestral plant form of secretory phospholipase A2, which through gene duplication and subsequent diversification, gave rise to the sPLA2-α clade during the plant colonization of earth but was secondarily lost in bryophytes and lycophytes; (ii) both clades were present already in the common ancestor of chlorophyte and streptophyte algae and were sometimes lost in particular groups; (iii) individual sPLA2 clades were subject to the independent horizontal gene transfer events during the evolution of separate Viridiplantae groups. These scenarios are not necessarily mutually exclusive, especially considering the widespread occurrence of horizontal gene transfer in green algae (Ma et al., 2022).
One notable element of plant sPLA2 evolution is the diversity in gene copy numbers for the two clades. The terminal duplication and diversification events of sPLA2 may be attributed to whole-genome duplication (eg. in some bryophyte species), polyploidization or amphidiploidization events (eg. N. tabacum or Z. mays). In addition, in some pteridophyte and gymnosperm species, sPLA2 underwent massive multiplication. Several hypotheses explain plant gene family expansion besides the whole-genome duplications and hybridization events. Gene families can expand through either segmental, tandem, or retro-transposition (RT) mechanism, but segmental and tandem duplication events were more predominant than RT (Kondrashov, 2012; Panchy et al., 2016). These new paralogs may perform an existing gene function (sub-functionalization) or acquire a novel role (neo-functionalization) (Panchy et al., 2016). These gene duplication mechanisms may often co-occur, as evidenced in the Eucalyptus grandis, which genome (n=11) has been shaped by lineage-specific genome duplication events and a high rate of tandem gene duplication (Myburg et al., 2014), leading to the highest number of sPLA2 members found in the analyzed angiosperms. As noted above, most of these within-family multiplication events occurred in the sPLA2-α clade, which is evolving considerably faster than the sPLA2-β clade. The notable exception is Arabidopsis (and the whole Brassicaceae clade, Supplementary Table 5, Supplementary Figure 5), where the expansion occurred predominantly in the sPLA2-β clade.
4.2 Is there an evolutionarily conserved role for the sPLA2-β clade in the male gametophyte endomembranes?
Our analysis of the sPLA2 expression revealed that while both -α and -β clades are present in various sporophytic tissues, sPLA2-α genes seem to be absent in the male gametophyte in dicots, monocots and Basal angiosperms (Figure 3A). This contrasts with the expression of the angiosperm sPLA2-β clade members, expressed either exclusively in the pollen and pollen tubes (Arabidopsis sPLA2-γ and -δ), or showing the highest expression in the male gametophyte (sPLA2-β from Arabidopsis, tomato, monocots, and Amborella). Significantly, our high-throughput data analysis is corroborated by the RT-PCR analysis of tobacco sPLA2 members (Figure 3B) and by multiple reports on Arabidopsis sPLA2 members (Lee et al., 2005; Kim et al., 2011). Although the split between the two sPLA2 clades occurred before the emergence of sexual reproduction via pollen, the presence of only sPLA2-β ortholog already in Amborella pollen and pollen tube suggests that the β-clade is the major sPLA2 in all angiosperms. Indeed, Kim et al. (2011) showed that when all three Arabidopsis PLA2-β clade members were suppressed by RNA interference, pollen development and germination were severely affected. Moreover, lysophosphatidylethanolamine, the product of sPLA2 activity, likely plays a vital role in pollen germination and pollen tube growth (Kim et al., 2011). Significant changes in lysophospholipid levels, including plasma membrane lysophosphatidylethanolamine and lysophosphatidylcholine, were recently described during tobacco pollen germination (Serrano et al., 2022). These findings further corroborate the conserved role of sPLA2 in the male gametophyte. Interestingly, compared to other beta-clade members, Arabidopsis sPLA2-γ shows the highest expression in the growing pollen tubes, suggesting that the three different β-clade members in Brassicaceae may have distinct roles during pollen development, germination, and tube growth.
A feature reportedly distinguishing plant sPLA2-α and -β clades is their distinct localization, suggesting that sPLA2-β may act primarily inside the endomembrane system and are not secreted to the apoplast (Fujikawa et al., 2012);. Three Arabidopsis sPLA2-β clade paralogs (β, γ, and δ) localized mainly to the ER and/or Golgi (Seo et al., 2008; Kim et al., 2011) while sPLA2-α localizes either to the apoplast or to the nucleus, depending on the plant status (Froidure et al., 2010; Jung et al., 2012). However, a canonical C-terminal ER-retention signal (KxEL) can be found only in 14 angiosperm sPLA2-β members and is missing even in Arabidopsis sPLA2-γ, and -δ. Therefore, the subcellular localization of sPLA2s, particularly the β-clade, might be variable and needs to be determined in other angiosperms.
4.3 The widespread presence of novel, evolutionary-constrained, PLA2-like gene family in plants
While annotating the plant’s sPLA2 sequences, we came across a subfamily of proteins with unusually long sequences (~250 aa), that possess a conserved PA2c domain containing both Ca2+ binding motif and HD catalytic dyad but lacking a signal peptide, which we termed PLA2-like. While the presence of the PLA2-like subfamily was briefly noticed before (Gupta et al., 2017), its phylogenomic distribution and sequence-structural properties were not explored. Our genome-wide analysis demonstrated that at least one PLA2-like member is consistently present from charophyte algae to higher plants, highlighting an ancient origin. It should be noted that PLA2-like genes can also be found in chlorophyte algae, but their distribution is patchy, suggesting either frequent losses or horizontal gene transfer in the plant group. Phylogenetically, PLA2-like sequences clustered into one clade, representing the independent evolutionary history of PLA2-like members and separating XI-A and XI-B sPLA2s. As noted above, the low gene-copy number of PLA2-like genes is under strong selection, effectively keeping PLA2-like as a single-copy gene in most species. While the putative enzymatic activity of PLA2-like remains obscure, the single-copy status is typically linked with genes often involved in essential metabolic processes and/or the formation of macromolecular complexes. Interestingly, the N-termini of PLA2-like proteins show intrinsically disordered region features, which function in protein-, DNA-, or RNA- binding (Han et al., 2014).
4.4 The angiosperm-conserved involvement of sPLA2 and PLA2-like genes in stress responses and developmental processes
Our analysis of cis-acting elements in promoters of evolutionarily-distinct angiosperms species suggested that the transcription of both sPLA2 and PLA2-like genes is regulated by various biotic, abiotic, and developmental stimuli (Figure 4). We have reported different cis-elements (W-box, WRE2, and WUN motif) involved in biotic stress responses, and found several methyl-jasmonate responsive elements in the sPLA2 promoters, further suggesting their active involvement during plant reactions to biotic stresses. The transcriptional activation of sPLA2 genes after a pathogen attack was shown in diverse species such as Arabidopsis (Froidure et al., 2010) and grapevine (Laureano et al., 2018), supporting our in-silico analysis. Analogously, sPLA2 promoters have multiple abiotic stress-responsive, abscisic acid-responsive and ethylene-responsive elements in the promoter region, suggesting that sPLA2 may have some regulatory roles during abiotic stresses. Verlotta et al., 2013 characterized durum wheat sPLA2s and assessed their involvement in drought stress (Verlotta et al., 2013). Similarly, Rice sPLA2 also showed a differential expression pattern in response to abiotic stress (Singh et al., 2012).
Besides the involvement in the stress responses, several case studies from distinct species corroborate the conserved transcriptional regulation of sPLA2 after developmental cues (Figure 4). Arabidopsis sPLA2-β is upregulated by auxin and in the curved regions of the peduncle, which were undergoing the gravitropic response (Lee et al., 2003). In Citrus sinensis, the sPLA2-α and β genes expression and enzyme activity in leaves and fruits exhibited diurnal rhythmic changes and light regulation, which suggested that diurnal fluctuations in lipophilic second messengers are involved in the regulation of physiological functions (Liao and Burns, 2010).
Very few high-throughput studies described the transcriptional regulation of the Arabidopsis PLA2-like gene. Interestingly it was demonstrated among genes with expression changes between pollen germination and tube growth (Wang et al., 2008), and listed among genes involved in Arabidopsis acyl lipid metabolism (Beisson et al., 2003). In the present study, we have predicted that PLA2-like members exhibited ubiquitous expression throughout the vegetative and reproductive tissues. In addition, it has light, stress, and hormonal-responsive elements in the promoter region that may be involved in diverse regulatory mechanisms and cellular functions.
4.5 Structural determinants of plant sPLA2 and PLA2-like superfamily
The comparative analysis of structural models for diverse plant sPLA2 and PLA2-like members strongly suggested that there are only subtle differences in the catalytic sites among typical members of sPLA2-α and -β clades. All tested models displayed characteristic sPLA2 features, shared with the experimentally-determined structures of non-plant sPLA2s from groups I, II, X and also rice XI-B sPLA2- namely three α-helices, a short two-stranded β-sheet, and a conserved calcium-binding loop (Dijkstra et al., 1981; Holland et al., 1990; White et al., 1990; Jabeen et al., 2006; Guy et al., 2009). On the other hand, the total conformation (due to the flexibility of N-terminal and C-terminal regions) and charge distribution differ substantially between the two clades or between different phylogenetic groups. For example, the electrostatic charge differences would affect the membrane-binding and protein-binding properties and thus may impact the sPLA2 catalytic functions. Notably, surface charge differences affecting the interactions with negatively-charged phospholipids were also described for non-plant sPLA2 members from groups IIA and X (Quach et al., 2014).
Importantly, template-free modelling of the C-terminal half of PLA2-like proteins suggested a fold similar to canonical sPLA2 structures, including a calcium-binding loop and catalytic dyad. However, the striking difference between the sPLA2 and PLA2-like is the loss of the disulfide bridges, where only four out of twelve Cys residues - forming two disulfide bridges - are conserved in PLA2-like sequences. On the other hand, the two retained disulfide bridges (C140-C167, C166-C192) - which connect helix 1 with the calcium loop and the helices 1 and 2 together - structurally correspond to those that are vital for the PLA2 catalytic activity of porcine PLA2 (C29-C45, C44-C105, Zhu et al., 1995). Therefore, despite the lower number of disulfide bridges retained in PLA2-like structures, PLA2-like members may still possess an enzyme catalytic activity towards lipidic substrates analogous to canonical sPLA2 (Mansfeld, 2009). In addition to the phylogenomic survey of canonical sPLA2 genes, our study may also serve as a call for further enzymological and physiological characterization of the enigmatic PLA2-like gene subfamily.
5 Conclusions
The widespread distribution of canonical sPLA2 and an unexpected presence of novel PLA2-like genes throughout the plant kingdom reflects an ancient sPLA2 origin and possible early split into two separate clades. This points to the conserved functional importance of plant sPLA2. The diverse evolutionary dynamic among the two sPLA2 clades and PLA2-like clade calls for future functional studies, which will be required to shed light on the functional importance of non-Arabidopsis β-clade members in plant reproduction, as well as the molecular characterization of PLA2-like genes and their involvement in plant cell physiology.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MP conceived and guided the study. AAS and MP carried out the analyses. AAS and MP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Czech Science Foundation grant GA21-09254S to MP.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1118670/full#supplementary-material
Supplementary Figure 1 | Sequence alignment of PA2c domain from all analyzed PLA2 proteins - shown separately for sPLA2- α, β (A) and PLA2-like (B) sequences. Calcium binding motif (red box) and highly conserved catalytic dyad Histidine and aspartic acid (HD) (blue box) are highlighted.
Supplementary Figure 2 | Predicted models of Nicotiana tabacum sPLA2-α, β, and PLA2-like. (A) Cartoon representation of structures. (B) Distribution of electrostatic potential on the PLA2 surface.
Supplementary Figure 3 | Predicted models of Amborella trichopoda PLA2-α, β, and PLA2-like. (A) Cartoon representation of structures. (B) Distribution of electrostatic potential on the PLA2 surface.
Supplementary Figure 4 | The positional conservation of cysteine disulfide bridges among canonical sPLA2 members from the -α and -β clade and PLA2-like proteins. Cysteine residues participating in disulfide bridges are shown in red. Arabidopsis sPLA2 and PLA2-like structural models are shown alongside experimental structure of sPLA2-α from rice.
Supplementary Figure 5 | Phylogenetic analysis of selected Brassicaceae family sPLA2 members. Phylogenetic tree using Oryza sativa sPLA2 members as an outgroup was constructed using the Maximum likelihood (PhyML) with the bootstrap method. The tree analysis was performed using the webserver (https://www.Phylogeny.fr: “One Click” Mode) with default parameters. The scale bar indicates the rates of substitutions/site. Abbreviations: AT, Arabidopsis thaliana; AL, Arabidopsis lyrata; Ah, Arabidopsis halleri; Carub, Capsella rubella; Cagra, Capsella grandiflora; Brara, Brassica rapa; Bol, Brassica oleracea; Camar, Cekile maritima; Sp, Schrenkiella parvula.
Supplementary Table 1 | List of identified secretory PLA2-α, β, and PLA2-like sequences with their protein length and chromosome locations. (ND – No data, AA – Amino acids).
Supplementary Table 2 | Primers used for the RT-PCR analysis.
Supplementary Table 3 | Comparison between Nicotiana tabacum Phospholipase A2 members.
Supplementary Table 4 | Assessment of sPLA2 and PLA2 structural models.
Supplementary Table 5 | Analysis of sPLA2 gene family in several Brassicaceae species.
Supplementary Data Sheet 1 | Protein sequences of sPLA2 and PLA2-like analyzed in this study.
Supplementary Data Sheet 2 | Promoter sequences of selected sPLA2 and PLA2-like genes analyzed in this study.
References
Abrams, Z. B., Johnson, T. S., Huang, K., Payne, P. R. O., Coombes, K. (2019). A protocol to evaluate RNA sequencing normalization methods. BMC Bioinf. 20, 679. doi: 10.1186/s12859-019-3247-x
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373 (6557), 871–876. doi: 10.1126/science.abj8754
Bahn, S. C., Lee, H. Y., Kim, H. J., Ryu, S. B., Shin, J. S. (2003). Characterization of arabidopsis secretory phospholipase A2-gamma cDNA and its enzymatic properties. FEBS Lett. 553 (1-2), 113–118. doi: 10.1016/S0014-5793(03)00982-7
Balsinde, J., Balboa, M. ,. A. (2005). Cellular regulation and proposed biological functions of group VIA calcium-independent phospholipase A2 in activated cells. Cell. Signal. 17, 1052–1062. doi: 10.1016/j.cellsig.2005.03.002
Beisson, F., Koo, A. J., Ruuska, S., Schwender, J., Pollard, M., Thelen, J. J., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. a 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132 (2), 681–697. doi: 10.1104/pp.103.022988
Bosch, M., Cheung, A. Y., Hepler, P. K. (2005). Pectin Methylesterase, a Regulator of Pollen Tube Growth. Plant Physiol. 138, 1334–1346. doi: 10.1104/pp.105.059865
Chapman, K. D. (1998). Phospholipase activity during plant growth and development and in response to environmental stress. Trends Plant Sci. 3 (11), 419–426. doi: 10.1016/S1360-1385(98)01326-0
Chen, G., Greer, M. S., Weselake, R. J. (2013). Plant phospholipase a: advances in molecular biology, biochemistry, and cellular function. Biomol. concepts 4 (5), 527–532. doi: 10.1515/bmc-2013-0011
Creelman, R. A., Mullet, J. E. (1997). Oligosaccharins, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell 9 (7), 1211–1223. doi: 10.1105/tpc.9.7.1211
Crooks, G. E., Hon, G., Chandonia, J. M., Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14 (6), 1188–1190. doi: 10.1101/gr.849004
Dhondt, S., Geoffroy, P., Stelmach, B. A., Legrand, M., Heitz, T. (2000). Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 23 (4), 431–440. doi: 10.1046/j.1365-313x.2000.00802.x
Dijkstra, B. W., Kalk, K. H., Hol, W. G., Drenth, J. (1981). Structure of bovine pancreatic phospholipase A2 at 1.7 Å resolution. J. Mol. Biol. 147 (1), 97–123. doi: 10.1016/0022-2836(81)90081-4
Ellinger, D., Stingl, N., Kubigsteltig, I. I., Bals, T., Juenger, M., Pollmann, S., et al. (2010). DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 153 (1), 114–127. doi: 10.1104/pp.110.155093
Froidure, S., Canonne, J., Daniel, X., Jauneau, A., Brière, C., Roby, D., et al. (2010). AtsPLA2-alpha nuclear relocalization by the arabidopsis transcription factor AtMYB30 leads to repression of the plant defence response. P. Natl. Acad. Sci. U.S.A. 107 (34), 15281–15286. doi: 10.1073/pnas.1009056107
Fujikawa, R., Fujikawa, Y., Iijima, N., Esaka, M. (2005). Molecular cloning, expression, and characterization of secretory phospholipase A2 in tobacco. Lipids 40 (9), 901–908. doi: 10.1007/s11745-005-1450-9
Fujikawa, Y., Fujikawa, Y., Iijima, N., Esaka, M. (2012). Characterization of secretory phospholipase A2 with phospholipase A1 activity in tobacco, Nicotiana tabacum (L.). Lipids 47 (3), 303–312. doi: 10.1007/s11745-011-3632-3
Gupta, P., Dash, P. K. (2017). Molecular details of secretory phospholipase A2 from flax (Linum usitatissimum l.) provide insight into its structure and function. Sci. Rep. 7 (1), 11080. doi: 10.1038/s41598-017-10969-9
Gupta, P., Saini, R., Dash, P. K. (2017). Origin and evolution of group XI secretory phospholipase A2 from flax (Linum usitatissimum) based on phylogenetic analysis of conserved domains. 3 Biotech. 7 (3), 1–10. doi: 10.1007/s13205-017-0790-x
Guy, J. E., Ståhl, U., Lindqvist, Y. (2009). Crystal structure of a class XIB phospholipase A2 (PLA2): rice (Oryza sativa) isoform-2 pla2 and an octanoate complex. J. Biol. Chem. 284 (29), 19371–19379. doi: 10.1074/jbc.M109.008466
Han, F., Peng, Y., Xu, L., Xiao, P. (2014). Identification, characterization, and utilization of single copy genes in 29 angiosperm genomes. BMC Genomics 15 (1), 504. doi: 10.1186/1471-2164-15-504
Holland, D. R., Clancy, L. L., Muchmore, S. W., Ryde, T. J., Einspahr, H. M., Finzel, B. C., et al. (1990). The crystal structure of a lysine 49 phospholipase A2 from the venom of the cottonmouth snake at 2.0-a resolution. J. Biol. Chem. 265 (29), 17649–17656. doi: 10.2210/pdb1ppa/pdb
Hyun, Y., Choi, S., Hwang, H. J., Yu, J., Nam, S. J., Ko, J., et al. (2008). Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 14 (2), 183–192. doi: 10.1016/j.devcel.2007.11.010
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13 (10), 2191–2209. doi: 10.1105/tpc.010192
Jabeen, T., Singh, N., Singh, R. K., Jasti, J., Sharma, S., Kaur, P., et al (2006). Crystal structure of a heterodimer of phospholipase A2 from Naja naja sagittifera at 2.3 A resolution reveals the presence of a new PLA2-like protein with a novel cys 32-Cys 49 disulphide bridge with a bound sugar at the substrate-binding site. Proteins 62, 329–337. doi: 10.1002/prot.20708
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi: 10.1038/s41586-021-03819-2
Jung, J., Kumar, K., Lee, H. Y., Park, Y. I., Cho, H. T., Ryu, S. B. (2012). Translocation of phospholipase A2α to apoplasts is modulated by developmental stages and bacterial infection in arabidopsis. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00126
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kato, T., Morita, M. T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M., et al. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of arabidopsis. Plant Cell 14 (1), 33–46. doi: 10.1105/tpc.010215
Kim, J. Y., Chung, Y. S., Ok, S. H., Lee, S. G., Chung, W. I., Kim, I. Y., et al. (1999). Characterization of the full-length sequences of phospholipase A2 induced during flower development. Biochim. Biophys. Acta 1489 (2-3), 389–392. doi: 10.1016/s0167-4781(99)00193-1
Kim, H. J., Ok, S. H., Bahn, S. C., Jang, J., Oh, S. A., Park, S. K., et al. (2011). Endoplasmic reticulum- and golgi-localized phospholipase A2 plays critical roles in arabidopsis pollen development and germination. Plant Cell 23 (1), 94–110. doi: 10.1105/tpc.110.074799
Kondrashov, F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279 (1749), 5048–5057. doi: 10.1098/rspb.2012.1108
La Camera, S., Balagué, C., Göbel, C., Geoffroy, P., Legrand, M., Feussner, I., et al. (2009). The arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol. Plant Microbe Interact. 22 (4), 469–481. doi: 10.1094/MPMI-22-4-0469
La Camera, S., Geoffroy, P., Samaha, H., Ndiaye, A., Rahim, G., Legrand, M., et al. (2005). A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in. Arabidopsis. Plant J. 44 (5), 810–825. doi: 10.1111/j.1365-313X.2005.02578.x
Laureano, G., Figueiredo, J., Cavaco, A. R., Duarte, B., Caçador, I., Malhó, R., et al. (2018). The interplay between membrane lipids and phospholipase a family members in grapevine resistance against. Plasmopara viticola. Sci. Rep. 8 (1), 1–15. doi: 10.1038/s41598-018-32559-z
Laxalt, A. M., Munnik, T. (2002). Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5 (4), 332–338. doi: 10.1016/s1369-5266(02)00268-6
Lee, H. Y., Bahn, S. C., Kang, Y. M., Lee, K. H., Kim, H. J., Noh, E. K., et al. (2003). Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in arabidopsis. Plant Cell 15 (9), 1990–2002. doi: 10.1105/tpc.014423
Lee, H. Y., Bahn, S. C., Shin, J. S., Hwang, I., Back, K., Doelling, J. H., et al. (2005). Multiple forms of secretory phospholipase A2 in plants. Prog. Lipid Res. 44 (1), 52–67. doi: 10.1016/j.plipres.2004.10.002
Lee, O. R., Kim, S. J., Kim, H. J., Hong, J. K., Ryu, S. B., Lee, S. H., et al. (2010). Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the arabidopsis root. Plant Cell 22 (6), 1812–1825. doi: 10.1105/tpc.110.074211
Lee, S., Suh, S., Kim, S., Crain, R. C., Kwak, M. J., Nam, H. G., et al. (1997). Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 12 (3), 547–556. doi: 10.1046/j.1365-313x.1997.00547.x
Liao, H.-L., Burns, J. K. (2010). Light controls phospholipase A2α and β gene expression in citrus sinensis. J. Exp. Bot. 61 (9), 2469–2478. doi: 10.1093/jxb/erq083
Li, M., Bahn, S. C., Fan, C., Li, J., Phan, T., Ortiz, M., et al. (2013). Patatin-related phospholipase pPLAIIIδ increases seed oil content with long-chain fatty acids in arabidopsis. Plant Physiol. 162 (1), 39–51. doi: 10.1104/pp.113.216994
Li, M., Bahn, S. C., Guo, L., Musgrave, W., Berg, H., Welti, R., et al. (2011). Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in arabidopsis. Plant Cell 23 (3), 1107–1123. doi: 10.1105/tpc.110.081240
Lo, M., Taylor, C., Wang, L., Nowack, L., Wang, T. W., Thompson, J. (2004). Characterization of an ultraviolet b-induced lipase in arabidopsis. Plant Physiol. 135 (2), 947–958. doi: 10.1104/pp.103.036376
Mansfeld, J. (2009). Plant phospholipases A2: perspectives on biotechnological applications. Biotechnol. Lett. 31 (9), 1373–1380. doi: 10.1007/s10529-009-0034-1
Mariani, M. E., Fidelio, G. D. (2019). Secretory phospholipases A2 in plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00861
Ma, J., Wang, S., Zhu, X., Sun, G., Chang, G., Li, L., et al. (2022). Major episodes of horizontal gene transfer drove the evolution of land plants. Mol. Plant 15 (5), 857–871. doi: 10.1016/j.molp.2022.02.001
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa015
Mueller, L. A., Solow, T. H., Taylor, N., Skwarecki, B., Buels, R., Binns, J., et al. (2005). The SOL genomics network: a comparative resource for solanaceae biology and beyond. Plant Physiol. 138 (3), 1310–1317. doi: 10.1104/pp.105.060707
Myburg, A. A., Grattapaglia, D., Tuskan, G. A., Hellsten, U., Hayes, R. D., Grimwood, J., et al. (2014). The genome of eucalyptus grandis. Nature 510 (7505), 356–362. doi: 10.1038/nature13308
One Thousand Plant Transcriptomes Initiative (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574 (7780), 679–685. doi: 10.1038/s41586-019-1693-2
Panchy, N., Lehti-Shiu, M., Shiu, S.-H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171 (4), 2294–2316. doi: 10.1104/pp.16.00523
Proost, S., Mutwil, M. (2018). CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res. 46 (W1), W133–W140. doi: 10.1093/nar/gky336
Quach, N. D., Arnold, R. D., Cummings, B. S. (2014). Secretory phospholipase A2 enzymes as pharmacological targets for treatment of disease. Biochem. Pharmacol. 90 (4), 338–348. doi: 10.1016/j.bcp.2014.05.022
Scherer, G. F. E. (2002). Secondary messengers and phospholipase A2 in auxin signal transduction. Plant Mol. Biol. 49, 357–372. doi: 10.1023/A:1015290510483
Seo, Y. S., Kim, E. Y., Kim, W. T. (2011). The arabidopsis sn-1-specific mitochondrial acylhydrolase AtDLAH is positively correlated with seed viability. J. Exp. Bot. 62 (15), 5683–5698. doi: 10.1093/jxb/err250
Seo, J., Lee, H. Y., Choi, H., Choi, Y., Lee, Y., Kim, Y. W., et al. (2008). Phospholipase A2β mediates light-induced stomatal opening in arabidopsis. J. Exp. Bot. 59 (13), 3587–3594. doi: 10.1093/jxb/ern208
Serrano, N., Pejchar, P., Soukupová, H., Hubálek, M., Potocký, M. (2022). Comprehensive analysis of glycerolipid dynamics during tobacco pollen germination and pollen tube growth. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1028311
Sibéril, Y., Doireau, P., Gantet, P. (2001). Plant bZIP G-box binding factors. modular structure and activation mechanisms. Eur. J. Biochem. 268 (22), 5655–5666. doi: 10.1046/j.0014-2956.2001.02552.x
Singh, A., Baranwal, V., Shankar, A., Kanwar, P., Ranjan, R., Yadav, S., et al. (2012). Rice phospholipase a superfamily: organization, phylogenetic and expression analysis during abiotic stresses and development. PloS One 7 (2), e30947. doi: 10.1371/journal.pone.0030947
Six, D. A., Dennis, E. A. (2000). The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim. Biophys. Acta 1488 (1-2), 1–19. doi: 10.1016/s1388-1981(00)00105-0
Ståhl, U., Ek, B., Stymne, S. (1998). Purification and characterization of a low-molecular-weight phospholipase A2 from developing seeds of Elm1. Plant Physiol. 117 (1), 197–205. doi: 10.1104/pp.117.1.197
Ståhl, U., Lee, M., Sjödahl, S., Archer, D., Cellini, F., Ek, B., et al. (1999). Plant low-molecular-weight phospholipase A2S (PLA2s) are structurally related to the animal secretory PLA2s and are present as a family of isoforms in rice (Oryza sativa). Plant Mol. Biol. 41 (4), 481–490. doi: 10.1023/a:1006323405788
Takáč, T., Novák, D., Šamaj, J. (2019). Recent advances in the cellular and developmental biology of phospholipases in plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00362
Unni, S., Huang, Y., Hanson, R. M., Tobias, M., Krishnan, S., Li, W. W., et al. (2011). Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 32 (7), 1488–1491. doi: 10.1002/jcc.21720
Verlotta, A., Liberatore, M. T., Cattivelli, L., Trono, D. (2013). Secretory phospholipases A2 in durum wheat (Triticum durum desf.): Gene expression, enzymatic activity, and relation to drought stress adaptation. Int. J. Mol. Sci. 14 (3), 5146–5169. doi: 10.3390/ijms14035146
Wang, S., Li, W., Liu, S., Xu, J. (2016). RaptorX-property: a web server for protein structure property prediction. Nucleic Acids Res. 44 (W1), W430–W435. doi: 10.1093/nar/gkw306
Wang, G., Ryu, S., Wang, X. (2012). Plant phospholipases: An overview. Methods Mol. Biol. 861, 123–137. doi: 10.1007/978-1-61779-600-5_8
Wang, Y., Zhang, W. Z., Song, L. F., Zou, J. J., Su, Z., Wu, W. H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in arabidopsis. Plant Physiol. 148 (3), 1201–1211. doi: 10.1104/pp.108.126375
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303. doi: 10.1093/nar/gky427
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., Barton, G. J. (2009). Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 (9), 1189–1191. doi: 10.1093/bioinformatics/btp033
White, S. P., Scott, D. L., Otwinowski, Z., Gelb, M. H., Sigler, P. B. (1990). Crystal structure of cobra-venom phospholipaseA2 in a complex with a transition-state analogue. Science 250 (4987), 1560–1563. doi: 10.1126/science.2274787
Yang, W., Devaiah, S. P., Pan, X., Isaac, G., Welti, R., Wang, X. (2007). AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and arabidopsis resistance to botrytis cinerea. J. Biol. Chem. 282 (25), 18116–18128. doi: 10.1074/jbc.M700405200
Yang, W. Y., Zheng, Y., Bahn, S. C., Pan, X. Q., Li, M. Y., Vu, H. S., et al. (2012). The patatin-containing phospholipase a pPLAIIα modulates oxylipin formation and water loss in Arabidopsis thaliana. Mol. Plant 5 (2), 452–460. doi: 10.1093/mp/ssr118
Keywords: sPLA2, plant, phylogeny, modelling, pollen, PLA2-like
Citation: Saddhe AA and Potocký M (2023) Comparative phylogenomic and structural analysis of canonical secretory PLA2 and novel PLA2-like family in plants. Front. Plant Sci. 14:1118670. doi: 10.3389/fpls.2023.1118670
Received: 07 December 2022; Accepted: 14 February 2023;
Published: 23 February 2023.
Edited by:
Eric Marechal, UMR5168 Laboratoire de Physiologie Cellulaire Vegetale (LPCV), FranceReviewed by:
Alexandre Noiriel, Université Claude Bernard Lyon 1, FranceHubert Schaller, UPR2357 Institut de biologie moléculaire des plantes (IBMP), France
Copyright © 2023 Saddhe and Potocký. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Potocký, potocky@ueb.cas.cz
 Ankush Ashok Saddhe
Ankush Ashok Saddhe Martin Potocký
Martin Potocký