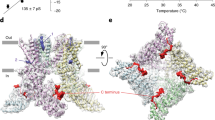
Abstract
Numerous physiological functions rely on distinguishing temperature through temperature-sensitive transient receptor potential channels (thermo-TRPs). Although the function of thermo-TRPs has been studied extensively, structural determination of their heat- and cold-activated states has remained a challenge. Here, we present cryo-EM structures of the nanodisc-reconstituted wild-type mouse TRPV3 in three distinct conformations: closed, heat-activated sensitized and open states. The heat-induced transformations of TRPV3 are accompanied by changes in the secondary structure of the S2-S3 linker and the N and C termini and represent a conformational wave that links these parts of the protein to a lipid occupying the vanilloid binding site. State-dependent differences in the behavior of bound lipids suggest their active role in thermo-TRP temperature-dependent gating. Our structural data, supported by physiological recordings and molecular dynamics simulations, provide an insight for understanding the molecular mechanism of temperature sensing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions of the paper are provided in the paper or the Supplementary Information. Cryo-EM density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession codes EMD-23853 (mTRPV3 closed, 4 °C, MSP2N2), EMD-23854 (mTRPV3 closed, 42 °C, MSP2N2), EMD-23855 (mTRPV3 sensitized, 42 °C, MSP2N2), EMD-23856 (mTRPV3 closed, 4 °C, cNW11), EMD-23857 (mTRPV3 closed, 42 °C, cNW11) and EMD-23858 (mTRPV3 open, 42 °C, cNW11) (Table 1). The corresponding model coordinates have been deposited to the Protein Data Bank (PDB) under accession codes 7MIJ (mTRPV3 closed, 4 °C, MSP2N2), 7MIK (mTRPV3 closed, 42 °C, MSP2N2), 7MIL (mTRPV3 sensitized, 42 °C, MSP2N2), 7MIM (mTRPV3 closed, 4 °C, cNW11), 7MIN (mTRPV3 closed, 42 °C, cNW11) and 7MIO (mTRPV3 open, 42 °C, cNW11) (Table 1). Source data are provided with this paper.
References
Castillo, K., Diaz-Franulic, I., Canan, J., Gonzalez-Nilo, F. & Latorre, R. Thermally activated TRP channels: molecular sensors for temperature detection. Phys. Biol. 15, 021001 (2018).
Voets, T. TRP channels and thermosensation. Handb. Exp. Pharmacol. 223, 729–741 (2014).
Islas, L. D. Thermal effects and sensitivity of biological membranes. Curr. Top. Membr. 74, 1–17 (2014).
Lamas, J. A., Rueda-Ruzafa, L. & Herrera-Pérez, S. Ion channels and thermosensitivity: TRP, TREK or both? Int. J. Mol. Sci. 20, 2371 (2019).
Arrigoni, C. & Minor, D. L. Global versus local mechanisms of temperature sensing in ion channels. Pflug. Arch. 470, 733–744 (2018).
Dhaka, A., Viswanath, V. & Patapoutian, A. Trp ion channels and temperature sensation. Annu. Rev. Neurosci. 29, 135–161 (2006).
Voets, T., Talavera, K., Owsianik, G. & Nilius, B. Sensing with TRP channels. Nat. Chem. Biol. 1, 85–92 (2005).
Latorre, R., Zaelzer, C. & Brauchi, S. Structure–functional intimacies of transient receptor potential channels. Q. Rev. Biophys. 42, 201–246 (2009).
Jordt, S. E., McKemy, D. D. & Julius, D. Lessons from peppers and peppermint: the molecular logic of thermosensation. Curr. Opin. Neurobiol. 13, 487–492 (2003).
Clapham, D. E. TRP channels as cellular sensors. Nature 426, 517–524 (2003).
Yuan, P. Structural biology of thermoTRPV channels. Cell Calcium 84, 102106 (2019).
Singh, A. K. et al. Structural basis of temperature sensation by the TRP channel TRPV3. Nat. Struct. Mol. Biol. 26, 994–998 (2019).
Nilius, B., Bíró, T. & Owsianik, G. TRPV3: time to decipher a poorly understood family member! J. Physiol. 592, 295–304 (2014).
Yang, P. & Zhu, M. X. Trpv3. Handb. Exp. Pharmacol. 222, 273–291 (2014).
Broad, L. M. et al. TRPV3 in drug development. Pharmaceuticals (Basel) 9, 55 (2016).
Chung, M. K., Lee, H., Mizuno, A., Suzuki, M. & Caterina, M. J. 2-Aminoethoxydiphenyl borate activates and sensitizes the heat-gated ion channel TRPV3. J. Neurosci. 24, 5177–5182 (2004).
Xiao, R. et al. Calcium plays a central role in the sensitization of TRPV3 channel to repetitive stimulations. J. Biol. Chem. 283, 6162–6174 (2008).
Liu, B., Yao, J., Zhu, M. X. & Qin, F. Hysteresis of gating underlines sensitization of TRPV3 channels. J. Gen. Physiol. 138, 509–520 (2011).
Moqrich, A. et al. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science 307, 1468–1472 (2005).
Xu, H. et al. TRPV3 is a calcium-permeable temperature-sensitive cation channel. Nature 418, 181–186 (2002).
Peier, A. M. et al. A heat-sensitive TRP channel expressed in keratinocytes. Science 296, 2046–2049 (2002).
Smith, G. D. et al. TRPV3 is a temperature-sensitive vanilloid receptor-like protein. Nature 418, 186–190 (2002).
Shimada, H. et al. The structure of lipid nanodisc-reconstituted TRPV3 reveals the gating mechanism. Nat. Struct. Mol. Biol. 27, 645–652 (2020).
Deng, Z. et al. Gating of human TRPV3 in a lipid bilayer. Nat. Struct. Mol. Biol. 27, 635–644 (2020).
Zubcevic, L., Borschel, W. F., Hsu, A. L., Borgnia, M. J. & Lee, S. Y. Regulatory switch at the cytoplasmic interface controls TRPV channel gating. Elife 8, e47746 (2019).
Zubcevic, L. et al. Conformational ensemble of the human TRPV3 ion channel. Nat. Commun. 9, 4773 (2018).
Singh, A. K., McGoldrick, L. L. & Sobolevsky, A. I. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat. Struct. Mol. Biol. 25, 805–813 (2018).
Nasr, M. L. et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 14, 49–52 (2017).
Shi, D. J., Ye, S., Cao, X., Zhang, R. & Wang, K. Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function. Protein Cell 4, 942–950 (2013).
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016).
Cao, E., Liao, M., Cheng, Y. & Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013).
Liu, B. & Qin, F. Single-residue molecular switch for high-temperature dependence of vanilloid receptor TRPV3. Proc. Natl Acad. Sci. USA 114, 1589–1594 (2017).
Cao, E., Cordero-Morales, J. F., Liu, B., Qin, F. & Julius, D. TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron 77, 667–679 (2013).
Feng, Q. Temperature sensing by thermal TRP channels: thermodynamic basis and molecular insights. Curr. Top. Membr. 74, 19–50 (2014).
Vlachová, V. et al. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J. Neurosci. 23, 1340–1350 (2003).
Yao, J., Liu, B. & Qin, F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl Acad. Sci. USA 108, 11109–11114 (2011).
Grandl, J. et al. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 13, 708–714 (2010).
Grandl, J. et al. Pore region of TRPV3 ion channel is specifically required for heat activation. Nat. Neurosci. 11, 1007–1013 (2008).
Brauchi, S., Orio, P. & Latorre, R. Clues to understanding cold sensation: thermodynamics and electrophysiological analysis of the cold receptor TRPM8. Proc. Natl Acad. Sci. USA 101, 15494–15499 (2004).
Brauchi, S., Orta, G., Salazar, M., Rosenmann, E. & Latorre, R. A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J. Neurosci. 26, 4835–4840 (2006).
Zhang, F. et al. Heat activation is intrinsic to the pore domain of TRPV1. Proc. Natl Acad. Sci. USA 115, E317–E324 (2018).
Voets, T. et al. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature 430, 748–754 (2004).
Gregorio-Teruel, L. et al. The integrity of the TRP domain is pivotal for correct TRPV1 channel gating. Biophys. J. 109, 529–541 (2015).
Kim, S. E., Patapoutian, A. & Grandl, J. Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS ONE 8, e59593 (2013).
Macikova, L., Vyklicka, L., Barvik, I., Sobolevsky, A. I. & Vlachova, V. Cytoplasmic inter-subunit interface controls use-dependence of thermal activation of TRPV3 channel. Int. J. Mol. Sci. 20, 3990 (2019).
Laursen, W. J., Schneider, E. R., Merriman, D. K., Bagriantsev, S. N. & Gracheva, E. O. Low-cost functional plasticity of TRPV1 supports heat tolerance in squirrels and camels. Proc. Natl Acad. Sci. USA 113, 11342–11347 (2016).
Clapham, D. E. & Miller, C. A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc. Natl Acad. Sci. USA 108, 19492–19497 (2011).
Jara-Oseguera, A. & Islas, L. D. The role of allosteric coupling on thermal activation of thermo-TRP channels. Biophys. J. 104, 2160–2169 (2013).
Kasimova, M. A. et al. A hypothetical molecular mechanism for TRPV1 activation that invokes rotation of an S6 asparagine. J. Gen. Physiol. 150, 1554–1566 (2018).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Russo, C. J. & Passmore, L. A. Electron microscopy: ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Hattori, M., Hibbs, R. E. & Gouaux, E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure 20, 1293–1299 (2012).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators and developers. Protein Sci. 30, 70–82 (2021).
Terwilliger, T. C., Ludtke, S. J., Read, R. J., Adams, P. D. & Afonine, P. V. Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927 (2020).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Dawaliby, R. et al. Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 291, 3658–3667 (2016).
Abraham, M. J. & Gready, J. E. Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J. Comput. Chem. 32, 2031–2040 (2011).
Lindorff-Larsen, K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010).
Jorgensen, W. L. & Tirado-Rives, J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc. Natl Acad. Sci. USA 102, 6665–6670 (2005).
Dittert, I. et al. Improved superfusion technique for rapid cooling or heating of cultured cells under patch-clamp conditions. J. Neurosci. Methods 151, 178–185 (2006).
Cao, C., Zakharian, E., Borbiro, I. & Rohacs, T. Interplay between calmodulin and phosphatidylinositol 4,5-bisphosphate in Ca2+-induced inactivation of transient receptor potential vanilloid 6 channels. J. Biol. Chem. 288, 5278–5290 (2013).
Luo, J., Stewart, R., Berdeaux, R. & Hu, H. Tonic inhibition of TRPV3 by Mg2+ in mouse epidermal keratinocytes. J. Invest. Dermatol. 132, 2158–2165 (2012).
Cao, X., Yang, F., Zheng, J. & Wang, K. Intracellular proton-mediated activation of TRPV3 channels accounts for the exfoliation effect of α-hydroxyl acids on keratinocytes. J. Biol. Chem. 287, 25905–25916 (2012).
Wang, H. et al. Mechanisms of proton inhibition and sensitization of the cation channel TRPV3. J. Gen. Physiol. 153, e202012663 (2021).
Choi, S. W. et al. The novel high-frequency variant of TRPV3 p.A628T in East Asians showing faster sensitization in response to chemical agonists. Pflug. Arch. 471, 1273–1289 (2019).
Doerner, J. F., Hatt, H. & Ramsey, I. S. Voltage- and temperature-dependent activation of TRPV3 channels is potentiated by receptor-mediated PI(4,5)P2 hydrolysis. J. Gen. Physiol. 137, 271–288 (2011).
Schrapers, K. T., Sponder, G., Liebe, F., Liebe, H. & Stumpff, F. The bovine TRPV3 as a pathway for the uptake of Na+, Ca2+ and NH4+. PLoS ONE 13, e0193519 (2018).
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Samsom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Acknowledgements
We thank S. Mulligan (Pacific Northwest Center for Cryo-EM), U. Baxa and T. Edwards (National Cancer Institute, The Frederick National Laboratory), R. Grassucci, Y.-C. Chi, Z. Zhang, C. Wang, J. Wang (Columbia University Cryo-Electron Microscopy Center) and H. Kuang (New York Structural Biology Center/National Center for CryoEM Access and Training) for help with microscope operation and data collection, personnel of the Supercomputer Center ‘Polytechnical’ at the St Petersburg Polytechnic University for access to the facility and D. Nolde for assistance with supercomputing. This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract no. HSSN261200800001E. Some of this work was performed at the Columbia University Cryo-Electron Microscopy Center. A portion of this research was supported by NIH grant no. U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Some of this work was performed at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and NY State Assembly Majority. MD simulations work was supported by the Russian Science Foundation (project #18-14-00375). Water and ion conductivity analysis was supported by the Russian Foundation for Basic Research (project #19-04-00350). Supercomputer calculations were supported within the framework of the HSE University Basic Research Program and the Russian Academic Excellence Project ‘5-100’. Whole-cell patch-clamp recordings were supported by Czech Science Foundation (project #19-03777 S). A.I.S. was supported by the NIH (R01 CA206573, R01 NS083660, R01 NS107253) and NSF (1818086).
Author information
Authors and Affiliations
Contributions
K.D.N. and A.N. made constructs, prepared protein samples and developed and optimized heating protocols. K.D.N., A.N. and N.K. performed cryo-EM data processing. K.D.N., A.N. and A.I.S. analyzed structural data. N.A.K., Y.A.T. and R.G.E. designed computational work, performed molecular modeling and analyzed the data. V.S. and V.V. prepared constructs and performed whole-cell patch-clamp recordings. E.Z. performed and analyzed planar lipid bilayer recordings. K.D.N., A.N., Y.A.T., V.V., E.Z., R.G.E. and A.I.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Chia-Hsueh Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Temperature-dependent changes in currents and thermodynamics of TRPV3.
a, A representative continuous recording of current from multiple TRPV3 channels occasionally reconstituted into the synthetic lipid bilayer (black) in response to the temperature ramp from 22 to 42 °C (red), with the membrane potential alternating between +100 mV (red circles) and –100 mV (blue circles). Note the sharp increase in current activity in the 36–42 °C temperature range. b, Temperature dependence of the open probability Po at +100 mV calculated using single-channel recordings (see examples in Fig. 1b; n = 19 independent experiments; 189,976 events were analyzed; the data includes the previously published12 and new recordings). Fitting of the data allows estimation of the temperature coefficient, Q10 = 27.0 ± 7.4 (n = 19 independent experiments). c, Van’t Hoff plot of the equilibrium constant Keq calculated using the values of Po (see Methods). Linear fits in the 22–36 °C and 36–42 °C temperature ranges provide the values of changes in enthalpy and entropy for the temperature-induced activation of TRPV3. Data in b and c are presented as mean values ± SEM. Source data for b and c are available online.
Extended Data Fig. 3 Map versus model FSC curves.
Map versus model FSC curves with and without mask were calculated using Mtriage as part of Phenix package59.
Extended Data Fig. 4 Cryo-EM density of TRPV3.
a, Stereo view of an ARD fragment of the 1.98-Å resolution cryo-EM map of TRPV3 reconstituted in MSP2N2 nanodiscs and incubated at 4 °C. b, Fragments of the same map for the membrane segments and TRP helix.
Extended Data Fig. 5 Comparison of pore geometry and architecture of TRPV3 structures.
a, Pore-forming domains of TRPV3 in the sensitized state with the residues lining the pore shown as sticks. Only two of four subunits are shown, with the front and back subunits omitted for clarity. The pore profile is shown as a space-filling model (light blue). b, Distribution of water (blue mesh) and ions (red mesh) during MD simulations of TRPV3 in the sensitized state, illustrating the lack of pore permeation. c, The pore radius for different TRPV3 structures calculated using HOLE79. The vertical dashed line denotes the radius of a water molecule, 1.4 Å. d, Superposition of a single subunit from different closed-state structures of TRPV3, with the main structural elements labeled. The root-mean-square deviation (RMSD) calculated for each pair of these subunits ranges between 0.314 and 0.441 Å. e-f, Superposition of TRPV3 structures in the closed (MSP2N2, 4 °C; green) and sensitized (pink) states (RMSD, 3.098 Å) viewed parallel to the membrane (e) or intracellularly (f). g-h, Superposition of TRPV3 structures in the closed (MSP2N2, 4 °C; green) and open (orange) states (RMSD, 3.293 Å) viewed parallel to the membrane (g) or intracellularly (h). Only two of four subunits are shown in (e) and (g) with the front and back subunits omitted for clarity. Note (e-h) that TRPV3 in the sensitized and open states becomes shorter and its intracellular skirt undergoes a clockwise rotation when viewed intracellularly.
Extended Data Fig. 6 Overview of cryo-EM data collected for mTRPV3 in cNW11 nanodiscs at 42 °C and 3D reconstruction workflow.
Representative micrographs with example particles circled in yellow and reference-free 2D class averages in different orientations are shown. Three datasets were collected and joined after particle clean-up. All processing steps were done in Relion, except the 2D/3D particle clean-up that was done in cryoSPARC.
Extended Data Fig. 7 Molecular dynamics simulations.
a, Conductance of water and Na+ ions through the selectivity filter and gate of the closed, sensitized and open TRPV3 plotted against the time course of MD simulation. Note that the closed state shows no permeation, the sensitized state permeates water through the selectivity filter only and the open state permeates water and Na+ ions through both selectivity filter and gate. b-g, Averaged MD density distributions (yellow) for non-hydrogen atoms of phosphatidylethanolamine (PE, b), phosphatidylinositol (PI, c), phosphatidylserine (PS, d), phosphatidylcholine (PC, e), phosphatidylglycerol (PG, f) and cholesterol (g) lipids nested in the vanilloid site of the closed TRPV3 at the beginning of 500-ns simulations. Black mesh shows cryo-EM density. MD snapshots of lipid molecules and residues coordinating their heads are shown in sticks. Chemical structures of the lipid molecules are shown to the left of each structural panel. The overlap of MD and EM densities are 26% for PE, 30% for PI, 36% for PS, 30% for PC, 33% for PG and 24% for cholesterol, calculated by multiplying the overlapping volume of MD and cryo-EM densities by two and dividing by their sum.
Extended Data Fig. 8 Comparison of open-state structures of wild-type TRPV3 and Y564A mutant.
a-b, Overall superposition (RMSD, 2.131 Å) of the open-state structures of wild-type TRPV3 (orange) and previously published Y564A mutant12 (blue, PDB ID: 6PVP) viewed parallel to the membrane (a) and extracellularly (b). c, Single-subunit superposition based on the transmembrane domains (RMSD, 0.943 Å). Note, the most pronounced conformational differences are observed for the S1-S2, S2-S3, S5-P and ARD loops, while the transmembrane domains and TRP helices superpose closely.
Extended Data Fig. 9 Sequence alignment of mouse TRPV channels.
α helices and β strands are depicted above the sequences as cylinders and arrows, respectively. The * symbols indicate residues in the ARD and linker domain that interact with residues in the C-terminus (¥ symbols). Red rectangular outlines denote regions involved in the interaction of the C-terminus with the ARD and linker domain, including residues conserved in thermo-TRPVs, and the AR5 and linker domain loops, which are present in thermo-TRPVs and absent in TRPV5–6. The location of the selectivity filter (S.F.) is indicated by a red box. Identical residues are colored red and highlighted in light pink. Positions of the previously identified mutations in TRPV3 that are critical for thermal sensitivity are highlighted in dark pink.
Extended Data Fig. 10 Conformational changes accompanying temperature-induced opening of wild-type TRPV3.
Superposition of the closed- and heat-activated open-state structures of TRPV3 (cNW11, 42 °C) viewed parallel to the membrane is shown in the centre. Insets show select regions with the arrows indicating the displacement of domains in the open relative to the closed state. The lipid at the vanilloid site is shown in sticks (pink).
Supplementary information
Supplementary Video 1
Structural heterogeneity of particles in a cryo-EM sample subjected to temperature cycles. 3D variability analysis of TRPV3 particles in cNW11 nanodiscs. Morph transition between open and closed states was calculated in cryoSPARC.
Supplementary Video 2
Structural transitions between closed, sensitized and open states. Regions involved in gating dynamics are highlighted in pink, with the elements undergoing the strongest structural changes highlighted in dark pink.
Source data
Source Data Extended Data Fig. 1
Statistical source data.
Rights and permissions
About this article
Cite this article
Nadezhdin, K.D., Neuberger, A., Trofimov, Y.A. et al. Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel. Nat Struct Mol Biol 28, 564–572 (2021). https://doi.org/10.1038/s41594-021-00615-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00615-4
This article is cited by
-
Looking back at 30 years of Nature Structural & Molecular Biology
Nature Structural & Molecular Biology (2024)
-
Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands
Nature Chemical Biology (2023)
-
Structure of human TRPV4 in complex with GTPase RhoA
Nature Communications (2023)
-
Thermoring basis for the TRPV3 bio-thermometer
Scientific Reports (2023)
-
Human TRPV1 structure and inhibition by the analgesic SB-366791
Nature Communications (2023)