Abstract
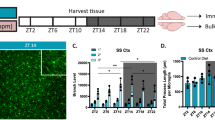
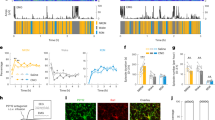
The activity of the immune system is controlled by circadian clocks present in different immune cells. The brain-resident subtype of immune cells, microglia, exhibits a wide range of functional phenotypes depending on the signaling molecules in their microenvironment. The exact role of microglia in the hypothalamic suprachiasmatic nuclei (SCN), the central circadian clock, has not been known. Therefore, the aim of this study was to determine (1) whether microenvironment-induced changes in microglial polarization affect circadian clocks in these cells and (2) whether the presence of microglia contributes to SCN clock function. Microglial and SCN clocks were monitored using PER2-driven bioluminescence rhythms at the tissue and single-cell levels. We found that polarization of resting microglia to a pro-inflammatory (M1) or anti-inflammatory (M2) state significantly altered the period and amplitude of their molecular circadian clock; importantly, the parameters changed plastically with the repolarization of microglia. This effect was reflected in specific modulations of the expression profiles of individual clock genes in the polarized microglia. Depletion of microglia significantly reduced the amplitude of the SCN clock, and co-cultivation of the SCN explants with M2-polarized microglia specifically improved the amplitude of the SCN clock. These results demonstrate that the presence of M2-polarized microglia has beneficial effects on SCN clock function. Our results provide new insight into the mutual interaction between immune and circadian systems in the brain.






Similar content being viewed by others
Data Availability
Python scripts used for image adjustment of bioluminescence microscopy recordings and bioluminescence data analyses are available at: https://github.com/clockgene. The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
References
Araki T, Ikegaya Y, Koyama R (2020) Microglia attenuate the kainic acid-induced death of hippocampal neurons in slice cultures. Neuropsychopharmacol Rep 40:85–91. https://doi.org/10.1002/npr2.12086
Barahona RA, Morabito S, Swarup V, Green KN (2022) Cortical diurnal rhythms remain intact with microglial depletion. Sci Rep 12:1–9. https://doi.org/10.1038/s41598-021-04079-w
Bartness TJ, Song CK, Demas GE (2001) SCN efferents to peripheral tissues: implications for biological rhythms. J Biol Rhythms 16:196–204. https://doi.org/10.1177/074873040101600302
Biber K, Neumann H, Inoue K, Boddeke HWGM (2007) Neuronal ‘On’ and ‘Off’ signals control microglia. Trends Neurosci 30:596–602. https://doi.org/10.1016/j.tins.2007.08.007
Blasko I, Stampfer-Kountchev M, Robatscher P et al (2004) How chronic inflammation can affect the brain and support the development of Alzheimer’s disease in old age: the role of microglia and astrocytes. Aging Cell 3:169–176. https://doi.org/10.1111/j.1474-9728.2004.00101.x
Bollinger T, Schibler U (2014) Circadian rhythms-From genes to physiology and disease. Swiss Med Wkly. https://doi.org/10.4414/smw.2014.13984
Bollinger T, Leutz A, Leliavski A et al (2011) Circadian clocks in mouse and human CD4+ T cells. PLoS ONE 6:e29801. https://doi.org/10.1371/journal.pone.0029801
Chen S, Fuller KK, Dunlap JC, Loros JJ (2020) A pro- and anti-inflammatory axis modulates the macrophage circadian clock. Front Immunol 11:867. https://doi.org/10.3389/fimmu.2020.00867
Cherry JD, Olschowka JA, O’Banion M (2014) Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J Neuroinflammation 11:98. https://doi.org/10.1186/1742-2094-11-98
Chhor V, Le Charpentier T, Lebon S et al (2013) Characterization of phenotype markers and neuronotoxic potential of polarised primary microglia in vitro. Brain Behav Immun 32:70–85. https://doi.org/10.1016/j.bbi.2013.02.005
Chi-Castañeda D, Ortega A (2016) Clock genes in glia cells. ASN Neuro 8:175909141667076. https://doi.org/10.1177/1759091416670766
Coleman LG, Zou J, Crews FT (2020) Microglial depletion and repopulation in brain slice culture normalizes sensitized proinflammatory signaling. J Neuroinflammation 17:27. https://doi.org/10.1186/s12974-019-1678-y
Cui L, Jin X, Xu F et al (2021) Circadian rhythm-associated Rev-erbα modulates polarization of decidual macrophage via the PI3K/Akt signaling pathway. Am J Reprod Immunol. https://doi.org/10.1111/aji.13436
Davalos D, Grutzendler J, Yang G et al (2005) ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8:752–758. https://doi.org/10.1038/nn1472
Duhart JM, Leone MJ, Paladino N et al (2013) Suprachiasmatic astrocytes modulate the circadian clock in response to TNF-. J Immunol 191:4656–4664. https://doi.org/10.4049/jimmunol.1300450
Durafourt BA, Moore CS, Zammit DA et al (2012) Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia 60:717–727. https://doi.org/10.1002/glia.22298
Fonken LK, Frank MG, Kitt MM et al (2015) Microglia inflammatory responses are controlled by an intrinsic circadian clock. Brain Behav Immun 45:171–179. https://doi.org/10.1016/j.bbi.2014.11.009
Gao Z, Zhu Q, Zhang Y et al (2013) Reciprocal modulation between microglia and astrocyte in reactive gliosis following the CNS injury. Mol Neurobiol 48:690–701. https://doi.org/10.1007/s12035-013-8460-4
Greenhalgh AD, David S (2014) Differences in the phagocytic response of microglia and peripheral macrophages after spinal cord injury and its effects on cell death. J Neurosci 34:6316–6322. https://doi.org/10.1523/JNEUROSCI.4912-13.2014
Greter M, Merad M (2013) Regulation of microglia development and homeostasis. Glia 61:121–127. https://doi.org/10.1002/glia.22408
Güldner FH (1983) Numbers of neurons and astroglial cells in the suprachiasmatic nucleus of male and female rats. Exp Brain Res 50:373–376. https://doi.org/10.1007/BF00239203
Hayashi Y (2013) Diurnal spatial rearrangement of microglial processes through the rhythmic expression of P2Y12 receptors. J Neurol Disord 01:1–7. https://doi.org/10.4172/2329-6895.1000120
Hu X, Leak RK, Shi Y et al (2014) Microglial and macrophage polarization—new prospects for brain repair. Nat Rev Neurol 11:56–64. https://doi.org/10.1038/nrneurol.2014.207
Ji K, Akgul G, Wollmuth LP, Tsirka SE (2013) Microglia actively regulate the number of functional synapses. PLoS ONE 8:e56293. https://doi.org/10.1371/journal.pone.0056293
Kabba JA, Xu Y, Christian H et al (2018) Microglia: housekeeper of the central nervous system. Cell Mol Neurobiol 38:53–71. https://doi.org/10.1007/s10571-017-0504-2
Kawai M, Rosen CJ (2010) PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol 6:629–636. https://doi.org/10.1038/nrendo.2010.155
Kettenmann H, Hoppe D, Gottmann K et al (1990) Cultured microglial cells have a distinct pattern of membrane channels different from peritoneal macrophages. J Neurosci Res 26:278–287. https://doi.org/10.1002/jnr.490260303
Lawson LJ, Perry VH, Gordon S (1992) Turnover of resident microglia in the normal adult mouse brain. Neuroscience 48:405–415. https://doi.org/10.1016/0306-4522(92)90500-2
Li Y, Yang Y-Y, Ren J-L et al (2017) Exosomes secreted by stem cells from human exfoliated deciduous teeth contribute to functional recovery after traumatic brain injury by shifting microglia M1/M2 polarization in rats. Stem Cell Res Ther 8:198. https://doi.org/10.1186/s13287-017-0648-5
Li Y, Shan Y, Desai RV et al (2020) Noise-driven cellular heterogeneity in circadian periodicity. Proc Natl Acad Sci 117:10350–10356. https://doi.org/10.1073/pnas.1922388117
Martínez-Tapia RJ, Chavarría A, Navarro L (2020) Differences in diurnal variation of immune responses in microglia and macrophages: review and perspectives. Cell Mol Neurobiol 40:301–309. https://doi.org/10.1007/s10571-019-00736-x
Matejuk A, Ransohoff RM (2020) Crosstalk between astrocytes and microglia: an overview. Front Immunol. https://doi.org/10.3389/fimmu.2020.01416
Mosser EA, Chiu CN, Tamai TK et al (2019) Identification of pathways that regulate circadian rhythms using a larval zebrafish small molecule screen. Sci Rep 9:12405. https://doi.org/10.1038/s41598-019-48914-7
Nakazato R, Hotta S, Yamada D et al (2017) The intrinsic microglial clock system regulates interleukin-6 expression. Glia 65:198–208. https://doi.org/10.1002/glia.23087
Norden DM, Trojanowski PJ, Walker FR, Godbout JP (2016) Insensitivity of astrocytes to interleukin 10 signaling following peripheral immune challenge results in prolonged microglial activation in the aged brain. Neurobiol Aging 44:22–41. https://doi.org/10.1016/j.neurobiolaging.2016.04.014
Novosadová Z, Polidarová L, Sládek M, Sumová A (2018) Alteration in glucose homeostasis and persistence of the pancreatic clock in aged mPer2Luc mice. Sci Rep 8:11668. https://doi.org/10.1038/s41598-018-30225-y
Okada K, Yano M, Doki Y et al (2008) Injection of LPS causes transient suppression of biological clock genes in rats. J Surg Res 145:5–12. https://doi.org/10.1016/j.jss.2007.01.010
Orihuela R, McPherson CA, Harry GJ (2016) Microglial M1/M2 polarization and metabolic states. Br J Pharmacol 173:649–665. https://doi.org/10.1111/bph.13139
Oster H, Damerow S, Kiessling S et al (2006) The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab 4:163–173. https://doi.org/10.1016/j.cmet.2006.07.002
Partch CL, Green CB, Takahashi JS (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24:90–99. https://doi.org/10.1016/j.tcb.2013.07.002
Peferoen L, Kipp M, van der Valk P et al (2014) Oligodendrocyte-microglia cross-talk in the central nervous system. Immunology 141:302–313. https://doi.org/10.1111/imm.12163
Pevet P, Challet E (2011) Melatonin: both master clock output and internal time-giver in the circadian clocks network. J Physiol 105:170–182. https://doi.org/10.1016/j.jphysparis.2011.07.001
Prinz M, Priller J (2014) Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci 15:300–312. https://doi.org/10.1038/nrn3722
Prolo LM, Takahashi JS, Herzog ED (2005) Circadian rhythm generation and entrainment in astrocytes. J Neurosci 25:404–408. https://doi.org/10.1523/JNEUROSCI.4133-04.2005
Rampersad SN (2012) Multiple applications of alamar blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 12:12347–12360. https://doi.org/10.3390/s120912347
Röhl C, Lucius R, Sievers J (2007) The effect of activated microglia on astrogliosis parameters in astrocyte cultures. Brain Res 1129:43–52. https://doi.org/10.1016/j.brainres.2006.10.057
Ronzano R, Roux T, Thetiot M et al (2021) Microglia-neuron interaction at nodes of Ranvier depends on neuronal activity through potassium release and contributes to remyelination. Nat Commun 12:5219. https://doi.org/10.1038/s41467-021-25486-7
Scheiermann C, Kunisaki Y, Frenette PS (2013) Circadian control of the immune system. Nat Rev Immunol 13:190–198. https://doi.org/10.1038/nri3386
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Silver AC, Arjona A, Hughes ME et al (2012) Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun 26:407–413. https://doi.org/10.1016/j.bbi.2011.10.001
Sládek M, Sumová A (2019) Modulation of NMDA-mediated clock resetting in the suprachiasmatic nuclei of mPer2Luc mouse by endocannabinoids. Front Physiol. https://doi.org/10.3389/fphys.2019.00361
Sládek M, Polidarová L, Nováková M et al (2012) Early chronotype and tissue-specific alterations of circadian clock function in spontaneously hypertensive rats. PLoS ONE 7:e46951. https://doi.org/10.1371/journal.pone.0046951
Sominsky L, Dangel T, Malik S et al (2021) Microglial ablation in rats disrupts the circadian system. FASEB J. https://doi.org/10.1096/fj.202001555RR
Son GH, Chung S, Choe HK et al (2008) Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci U S A 105:20970–20975. https://doi.org/10.1073/pnas.0806962106
Szepesi Z, Manouchehrian O, Bachiller S, Deierborg T (2018) Bidirectional microglia-neuron communication in health and disease. Front Cell Neurosci. https://doi.org/10.3389/fncel.2018.00323
Takahashi K, Yamamura F, Naito M (1989) Differentiation, maturation, and proliferation of macrophages in the mouse yolk sac: a light-microscopic, enzyme-cytochemical, immunohistochemical, and ultrastructural study. J Leukoc Biol 45:87–96. https://doi.org/10.1002/jlb.45.2.87
Tang Y, Le W (2016) Differential roles of M1 and M2 microglia in neurodegenerative diseases. Mol Neurobiol 53:1181–1194. https://doi.org/10.1007/s12035-014-9070-5
Tarassishin L, Suh HS, Lee SC (2014) LPS and IL-1 differentially activate mouse and human astrocytes: Role of CD14. Glia 62:999–1013. https://doi.org/10.1002/glia.22657
Timmons GA, O’Siorain JR, Kennedy OD et al (2020) Innate rhythms: clocks at the center of monocyte and macrophage function. Front Immunol. https://doi.org/10.3389/fimmu.2020.01743
Tinevez J-Y, Perry N, Schindelin J et al (2017) TrackMate: an open and extensible platform for single-particle tracking. Methods 115:80–90. https://doi.org/10.1016/j.ymeth.2016.09.016
Tomida M, Yamamoto-Yamaguchi Y, Hozumi M (1984) Purification of a factor inducing differentiation of mouse myeloid leukemic M1 cells from conditioned medium of mouse fibroblast L929 cells. J Biol Chem 259:10978–10982
Wang X-L, Wolff SEC, Korpel N et al (2020) Deficiency of the circadian clock gene Bmal1 reduces microglial immunometabolism. Front Immunol. https://doi.org/10.3389/fimmu.2020.586399
Wen L, You W, Wang H et al (2018) Polarization of microglia to the M2 phenotype in a peroxisome proliferator-activated receptor gamma-dependent manner attenuates axonal injury induced by traumatic brain injury in mice. J Neurotrauma 35:2330–2340. https://doi.org/10.1089/neu.2017.5540
Yamamoto T, Nakahata Y, Tanaka M et al (2005) Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem 280:42036–42043. https://doi.org/10.1074/jbc.M509600200
Yamazaki S, Takahashi JS (2005) In real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol 393:288–301
Yoo S-H, Yamazaki S, Lowrey PL et al (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101:5339–5346. https://doi.org/10.1073/pnas.0308709101
Acknowledgements
This work was supported by the Grant Agency of Charles University (project GA UK No. 514219), OPPK BrainView CZ.2.16/3.1.00/21544, and Research Project RV0 67985823. The authors thank Eva Tlusta for technical assistance. We would also like to thank Chun-Xia Yi and Irina Milanova (Amsterdam UMC, Amsterdam, Netherlands) for their support and methodology sharing.
Funding
This work was supported by the Grant Agency of Charles University (project GA UK No. 514219), OPPK BrainView CZ.2.16/3.1.00/21544, and Research Project RV0 67985823.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study execution. PH and AS contributed to study conception and design. PH and KS contributed to material preparation, data collection, and analysis. PH contributed to the first draft of the manuscript. AS reviewed and commented on the following versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
The Animal Care and Use Committee of the Institute of Physiology, in agreement with the Animal Protection Law of the Czech Republic as well as European Community Council directives 86/609/EEC, approved all experiments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (AVI 1392 KB) Online Resource 1 Bright-field video recording of M0-polarized microglia culture recorded for 96 hours. Snapshots were acquired every 3 hours.
Supplementary file2 (AVI 1210 KB) Online Resource 2 Bright-field video recording of M1-polarized microglia culture recorded for 96 hours. Snapshots were acquired every 3 hours.
Supplementary file3 (AVI 1516 KB) Online Resource 3 Bright-field video recording of M2-polarized microglia culture recorded for 96 hours. Snapshots were acquired every 3 hours.
Supplementary file4 (AVI 4536 KB) Online Resource 4 Bioluminescence video recording of M0-polarized microglia culture recorded for 96 hours. Snapshots were acquired every 3 hours.
Supplementary file5 (AVI 4883 KB) Online Resource 5 Bioluminescence video recording of M1-polarized microglia culture recorded for 96 hours. Snapshots were acquired every 3 hours.
Supplementary file6 (AVI 5009 KB) Online Resource 6 Bioluminescence video recording of M2-polarized microglia culture recorded for 96 hours. Snapshots were acquired every 3 hours.
10571_2022_1252_MOESM7_ESM.pdf
Supplementary file7 (PDF 756 KB) Online Resource 7 Supplemental Figure S1 summarizing obtained results regarding the effect of PPARγ activation/inhibition on PER2-driven bioluminescence rhythms in differently polarized microglia.
Rights and permissions
About this article
Cite this article
Honzlová, P., Semenovykh, K. & Sumová, A. The Circadian Clock of Polarized Microglia and Its Interaction with Mouse Brain Oscillators. Cell Mol Neurobiol 43, 1319–1333 (2023). https://doi.org/10.1007/s10571-022-01252-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-022-01252-1