-
PDF
- Split View
-
Views
-
Cite
Cite
Guillem Pascual-Aznar, Grzegorz Konert, Martina Bečková, Eva Kotabová, Zdenko Gardian, Jana Knoppová, Lenka Bučinská, Radek Kaňa, Roman Sobotka, Josef Komenda, Psb35 Protein Stabilizes the CP47 Assembly Module and Associated High-Light Inducible Proteins during the Biogenesis of Photosystem II in the Cyanobacterium Synechocystis sp. PCC6803, Plant and Cell Physiology, Volume 62, Issue 1, January 2021, Pages 178–190, https://doi.org/10.1093/pcp/pcaa148
Close - Share Icon Share
Abstract
Photosystem II (PSII) is a large membrane protein complex performing primary charge separation in oxygenic photosynthesis. The biogenesis of PSII is a complicated process that involves a coordinated linking of assembly modules in a precise order. Each such module consists of one large chlorophyll (Chl)-binding protein, number of small membrane polypeptides, pigments and other cofactors. We isolated the CP47 antenna module from the cyanobacterium Synechocystis sp. PCC 6803 and found that it contains a 11-kDa protein encoded by the ssl2148 gene. This protein was named Psb35 and its presence in the CP47 module was confirmed by the isolation of FLAG-tagged version of Psb35. Using this pulldown assay, we showed that the Psb35 remains attached to CP47 after the integration of CP47 into PSII complexes. However, the isolated Psb35-PSIIs were enriched with auxiliary PSII assembly factors like Psb27, Psb28-1, Psb28-2 and RubA while they lacked the lumenal proteins stabilizing the PSII oxygen-evolving complex. In addition, the Psb35 co-purified with a large unique complex of CP47 and photosystem I trimer. The absence of Psb35 led to a lower accumulation and decreased stability of the CP47 antenna module and associated high-light-inducible proteins but did not change the growth rate of the cyanobacterium under the variety of light regimes. Nevertheless, in comparison with WT, the Psb35-less mutant showed an accelerated pigment bleaching during prolonged dark incubation. The results suggest an involvement of Psb35 in the life cycle of cyanobacterial Chl-binding proteins, especially CP47.
Introduction
Photosystem II (PSII), localized in the thylakoid membranes (TMs) of plants, algae and cyanobacteria, carries out the highly energetically demanding reaction of water splitting. This intricate multi-subunit complex is the light-driven water–plastoquinone:oxidoreductase, responsible for the release of oxygen, protons and electrons from water molecules. In combination with Photosystem I (PSI) and the cytochrome b6f complex, PSII generates the ATP and NADPH that are used for the fixation of carbon dioxide during reactions of the Calvin–Benson cycle.
The detailed structure of dimeric PSII from cyanobacteria, now known to a resolution of 1.9 Å (Umena et al. 2011), has unveiled the spatial configuration of protein subunits and cofactors involved in excitation energy transfer and electron transport. At the heart of the complex lie the two reaction center (RC) subunits, named D1 and D2, that bind chlorophyll (Chl) and several other cofactors involved in primary charge separation. Two other Chl-binding subunits, CP47 and CP43, flank both sides of the RCII and serve as inner antenna proteins funneling excitation energy from the outer antenna toward the RCII. These four large subunits are accompanied by 13 small subunits, mostly containing the single transmembrane helix. The complex is shielded from the lumenal side by three extrinsic subunits called PsbO, PsbU and PsbV that lie in the vicinity of the manganese cluster (Umena et al. 2011).
In cyanobacteria, it is thought that biogenesis of PSII occurs at the periphery of the cell in layers of photosynthetic TMs adjacent to the plasma membrane (Selaõ et al. 2016, Rast et al. 2019). The process proceeds in a stepwise fashion by a combination of four assembly modules consisting of a large Chl-binding subunit (D1, D2, CP43 and CP47) with adjacent small subunits and pigments (Komenda et al. 2012b). The earliest assembly step is the formation of the PSII RC complex (RCII) from the D1 and D2 modules (D1m and D2m), followed by the attachment of the CP47 module (CP47m) giving rise to a PSII core complex lacking CP43 (termed RC47). After RC47 is made, the CP43 module (CP43m) is attached to form a monomeric PSII core complex. Final steps of assembly include the light-driven assembly of the manganese cluster on the lumenal side of PSII, the attachment of the extrinsic subunits and the dimerization of the complex to form the fully functional PSII dimer [PSII(2)] (Komenda et al. 2012b).
PSII assembly needs to be highly organized since the release of Chl could lead to photo-oxidative damage endangering cell survival. Therefore, this process requires the assistance of auxiliary proteins that are not found in the mature PSII complex but instead interact transiently with distinct PSII subunits and assembly modules. During the last decade, various PSII assembly protein factors have been identified and their association with particular assembly modules has been determined (Komenda et al. 2012b). Although for most of these protein factors we also know their detailed 3D structure (Cormann et al. 2009, Mabbitt et al. 2009, Michoux et al. 2010, Yang et al. 2011, Jackson et al. 2012, Michoux et al. 2012, Bialek et al. 2013, Michoux et al. 2014, Bečková et al. 2017b, Yu et al. 2018), the role of assembly factors is generally poorly characterized.
Apart from the CP43m, CP47m is the only PSII assembly module that has been purified and characterized (Boehm et al. 2011). In agreement with the detailed structural model of PSII (Umena et al. 2011), CP47m contains PsbH, PsbL and PsbT and bound Chls and β-carotene molecules. In addition, three members (HliA, HliB, HliC) of the family of high-light-inducible proteins (Hlips) associate with the CP47m (Promnares et al. 2006, Yao et al. 2007). Hlips are small, single transmembrane helix proteins that bind Chl and β-carotene (for review see Komenda and Sobotka 2016) and are considered to participate in excess energy quenching, Chl recycling and/or Chl delivery. Hlips remain attached to CP47m even after the binding of this module to the RCII complexes (Promnares et al. 2006, Boehm et al. 2012).
Interestingly, in two thermophilic cyanobacterial strains (Synechococcus OS-A′ and OS-B′) the Hlip domain has been found in a protein, which contains additional transmembrane helix (Kilian et al. 2008). The function of this second helix is unknown, but genomes of most cyanobacteria contain separate genes encoding a putative homologue of this protein part. In the model cyanobacterium Synechocystis PCC 6803 (hereafter Synechocystis), this homolog is encoded by the ssl2148 gene and is annotated as hypothetical protein with a putative single transmembrane helix.
In the present study, we characterized CP47m in more detail and identified its new protein component, which we named here Psb35 and which is encoded by the ssl2148 gene. The protein is also present in other PSII complexes containing CP47, but it is most probably missing in the active, fully assembled PSII. Psb35, possessing a long cytoplasmic C-terminus, increases the accumulation and stability of the CP47m and associated Hlips (HliA/B proteins). Probably as a consequence of the impaired CP47m stability, the Synechocystis Psb35-less mutant exhibits a faster loss of PSII and PSI complexes during the dark incubation, suggesting the importance of Psb35 for maintaining the cellular level of Chl in the dark.
Results
Psb35 is a membrane protein associates with the CP47m and larger CP47-containig PSII complexes
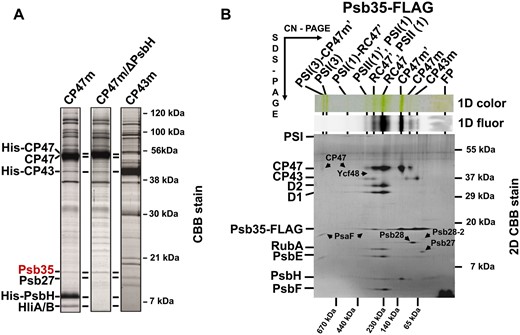
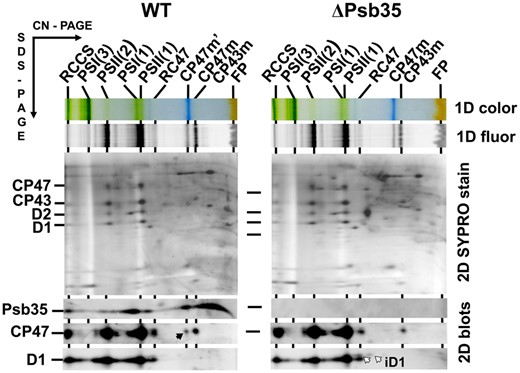
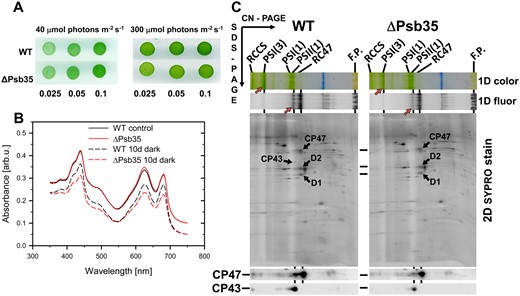
In our previous studies, we have characterized in detail the composition of both CP47m and CP43m (Boehm et al. 2011). Both modules, purified using a combination of nickel affinity and anion exchange chromatography, contained Chls, β-carotenes and most of small subunits identified in contact with the CP47 and CP43 proteins in the high-resolution crystallographic PSII model (Umena et al. 2011). Moreover, several auxiliary factors like Ycf48 (Yu et al. 2018), Pam68 (Bučinská et al. 2018) and Psb27 (Cormann et al. 2009, Mabbitt et al. 2009, Komenda et al. 2012a), which are absent in crystalized PSII, have also been detected in the isolated modules. Nevertheless, the combination of affinity and ion exchange chromatography used for the isolation of CP43m and CP47m could result in a loss of more weakly bound auxiliary factors. Additional factors associated with CP47m would be particularly interesting given the specific interaction of this module with Hlips (HliA/B proteins) and strict dependence of the CP47 synthesis on the availability of de novo produced Chl (Hollingshead et al. 2016). We therefore analyzed the protein composition of CP47m isolated using just a single Ni-affinity purification step. We used a previously described strain unable to assemble RCII due to the absence of cytochrome b-559 (Komenda et al. 2004), which expresses CP47-associated His-PsbH and also lacks FtsH2 protease to prevent the degradation of unassembled PSII assembly intermediates (see Supplementary Table S1, D'Haene et al. 2015). We compared the composition of CP47m with its variant lacking PsbH (CP47m/ΔPsbH) and also with CP43m isolated from the strain expressing His-CP43 and lacking the D1 protein (Boehm et al. 2011). The variant of CP47m lacking PsbH was investigated due to the previously observed effect of the protein on the binding of Hlips (Promnares et al. 2006) and Pam68 (Bučinská et al. 2018) to CP47. The strain used for the isolation of CP47m/ΔPsbH expresses His-CP47 and lacks PsbH and FtsH2 and it is unable to assemble RCII due to the absence of D1 (D'Haene et al. 2015). Mass spectrometric analysis of the protein bands, resolved in both CP47 modules using SDS-PAGE (Fig. 1A), showed the reproducible occurrence of a 11-kDa protein band that we identified as a hypothetical protein Ssl2148. This protein was absent in the affinity-purified CP43m (Fig. 1A) and also in a wild-type (WT) control elution (not shown). Due to its presence in the PSII assembly module, we named the protein Psb35.
Detection of the Psb35 (Ssl2148) protein in PSII assembly modules CP47m, CP47m lacking PsbH (CP47m/ΔPsbH) and CP43m (A), and 2D analysis of preparation isolated by a single-step FLAG-specific affinity chromatography from the strain expressing Psb35-FLAG instead of native Psb35 (B). (A) The modules were isolated by a single-step Ni-affinity chromatography and analyzed by SDS-PAGE. The gel was stained by Coomassie Blue and designated proteins were identified by MS. One microgram of Chl was loaded for each preparation. (B) The isolated preparation was analyzed by 2D CN/SDS-PAGE. After the first dimension, the gel was photographed (1D color) and scanned for Chl fluorescence (1D fluor). After the separation in the second dimension, the 2D gel was stained using Coomassie Blue (2D CBB stain) and the stained minor proteins designated by arrows were detected by MS. The identity of the major components was verified by Western blotting (see Supplementary Table S2). The designation of complexes: PSI(3) and PSI(1), trimeric and monomeric PSI; PSI(3)-CP47m′, PSI(3) with bound CP47m and Psb35; PSII(1), monomeric PSII; RC47, PSII(1) lacking CP43; PSI(1)-RC47, PSI(1) with bound RC47; complexes designated with a prime contain Psb35-FLAG; FP, free pigments. One microgram of Chl was loaded.
To further confirm the association of Psb35 with the CP47m, we used a newly constructed strain expressing the Psb35 protein with 3xFLAG tag attached to the C-terminus of the protein (Psb35-FLAG). The tagged Psb35 was used as bait and associated proteins were isolated from detergent-solubilized mutant membranes by a FLAG-specific immunoaffinity purification and analyzed by clear native/SDS gel electrophoresis (2D CN/SDS-PAGE) (Fig. 1B). Using this approach, we were able to pull down the CP47m but also larger complexes containing CP47, namely PSII monomer [PSII(1)], PSII monomer lacking CP43 (RC47) and also this complex associated with PSI monomer [PSI(1)-RC47]. The complexes containing Psb35 (designated by prime) were clearly detected in the gel along with their versions, which had lost this protein, most probably during the native electrophoresis. There was no apparent dimeric PSII complex.
Interestingly, the mass spectrometric analysis of protein spots resolved from the isolated Psb35-FLAG preparation by 2D CN/SDS-PAGE showed (Supplementary Table S2) the absence of lumenal proteins stabilizing the oxygen-evolving complex (Fig. 1B, for the composition of FLAG-tagged oxygen-evolving PSII, see Trinugroho et al. 2020). On the other hand, the preparation contained a number of PSII auxiliary factors that assist PSII biogenesis, such as RubA (Kiss et al. 2019), Psb27 (Komenda et al. 2012a), Psb28 (Dobáková et al. 2009), Psb28-2 (Bečková et al. 2017a) and Ycf48 (Yu et al. 2018). Based on the staining intensity of the spots, RubA and Psb28 were present in nearly stoichiometric amounts to the PSII core complexes. We also confirmed the interaction of Psb35 with the specific populations of PSII complexes associated with Psb28 and RubA by detecting the Psb35 in the preparations previously isolated using Psb28-FLAG and FLAG-RubA, respectively (Supplementary Figs. S1, S2, see also Bečková et al. 2017a, Kiss et al. 2019).
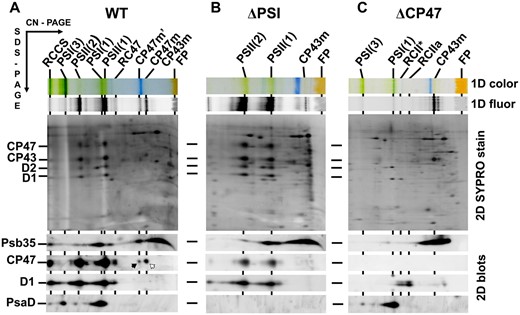
To further confirm the occurrence of Psb35 in CP47m, RC47 and PSII, we used a specific Psb35 antibody raised against the last 14-amino-acid residues of the protein to screen solubilized membrane complexes of WT for the presence of Psb35. Using 2D CN/SDS-PAGE of membrane proteins combined with immunobloting, we detected Psb35 among unassembled proteins but also in larger protein complexes (Fig. 2A). The parallel analysis of WT membranes along with membranes of the PSI-less (Fig. 2B) and CP47-less (Fig. 2C) mutants enabled to establish the presence of Psb35 in CP47 containing complexes identical to those detected in the Psb35-FLAG preparation. In addition, the protein was detected in a PSII–PSI supercomplex (RCCS) and in the dimeric PSII core complex [PSII(2)], which was especially apparent in the PSI-less strain (Fig. 2B) Interestingly, in the CP47-less background, Psb35 was absent in larger complexes except a low intensity band, which we assigned to a small population of a larger monomeric PSI complex (Fig. 2C) . In summary, the data confirmed the strictly CP47-dependent association of Psb35 with PSII complexes.
Identification of the Psb35 protein in the membranes of WT, PSI-less (ΔPSI) and CP47-less (ΔCP47) strains. Membranes isolated from the strains were analyzed by 2D CN/SDS-PAGE. After the first dimension, the gel was photographed (1D color) and scanned for Chl fluorescence (1D fluor). After the separation in the second dimension, the 2D gel was stained using Sypro Orange (2D SYPRO stain) and blotted to a poly(vinylidene fluoride) membrane (2D blots) and Psb35, CP47, D1 and PsaD were detected by the specific antibodies. The designation of complexes is as in Fig. 1. RCCS designates a supercomplex of PSI and PSII; PSII(2) dimeric PSII core complex. Black arrow designates CP47 within CP47m′, and the white arrow designates CP47 within CP47m. 5, 1.5 and 3 μg of Chl were loaded for WT, ΔPSI and ΔCP47 strains, respectively.
A supercomplex between trimeric PSI and CP47m
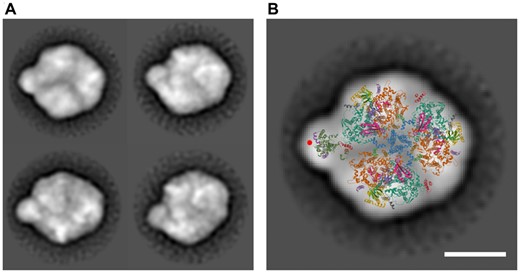
Among the isolated protein complexes containing Psb35-FLAG, the most interesting for us was the complex somewhat larger than the PSI trimer and containing PSI subunits, CP47 and Psb35-FLAG. We therefore ascribed it to a putative PSI trimer with bound CP47 [PSI(3)-CP47m′]. This complex could also be isolated from a strain expressing the His-tagged CP47 in the CP43-less background (Boehm et al. 2012) (Supplementary Fig. S3). When membranes isolated from this strain were subjected to the Ni-affinity chromatography, we obtained a preparation containing RC47, several forms of CP47m and the PSI(3)-CP47m′ complex as judged from the immunoblot signals of PsaF, CP47 and Psb35 detected in the green bands. The most abundant forms of CP47m were the standard CP47m and CP47m containing Psb35 (CP47m′) while the origin of larger and less intensive CP47m″ and CP47m‴ bands remains unknown. To confirm the identity of the PSI(3)-CP47m′ complex, we subjected it to an electron microscopic negative staining single-particle analysis. We used the complex eluted from the gel after native electrophoresis of the isolated His-tagged CP47 preparation. Processing of the images revealed several complex populations mostly differing in the angle under which the complex was sitting on the electron microscopic grid (Fig. 3A). The image of the most abundant species with the best resolution univocally documented that indeed the complex is the PSI trimer with CP47 partly inserted into the space between two monomers within the PSI trimer (Fig. 3B).
Electron microscopic analysis of the PSI(3)-CP47m complex co-isolated with His-CP47 and separated by CN PAGE. (A) The negatively stained particles of the complex (left panel) were obtained by the classification of 5,992, 1,205, 2,424 and 1,426 particles. The projection was overlaid with a cyanobacterial X-ray model of the PSI trimer and CP47 antenna with PsbH (right panel). CP47 is shown in dark green, PsbH in purple and a density putatively ascribed to the Psb35 is designated by a red circle. The coordinates are taken from Protein Data Bank (http://www.rcsb.org/pdb), PSI Code 1JB0 (Jordan et al. 2001) and PSII code 3WU2 (Umena et al. 2011). The scale bars represent 10 nm.
Membrane orientation of Psb35 and its cellular localization
The membrane orientation of Psb35 was analyzed in right-side out membrane vesicles isolated from WT (Komenda et al. 2002) before and after the treatment with trypsin. We also trypsinized the vesicles of strain lacking the original Psb35 but instead expressing Psb35-FLAG. While the untreated membranes of both WT and Psb35-FLAG strains contained specific bands, which reacted with the C-terminal-specific antibody and well corresponded with the size of Psb35 or its tagged version, after trypsinization both bands disappeared (Supplementary Fig. S4A). Since the arginine and lysine residues required for trypsin cleavage are abundant in the C-terminal protrusion but absent in the N-terminal tail, the data show that the long C-terminus of Psb35 is facing the cytoplasm while the short N-terminus is protruding on the lumenal side of the membranes (Supplementary Fig. S4B). The experiment was validated by monitoring the intactness of the control lumenal Ycf48 protein, which remained uncleaved during the assay.
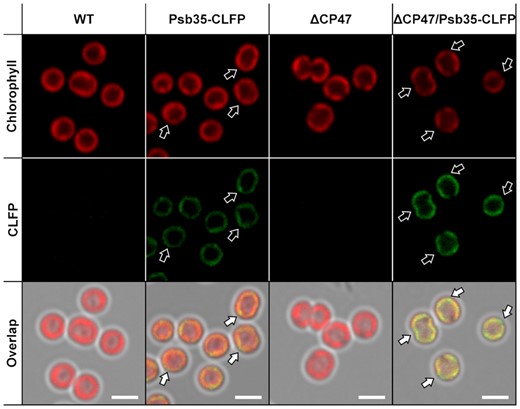
The localization of Psb35 protein at the cellular level was investigated using confocal microscope in the strain expressing mClover3, a derivative of Green Fluorescent Protein, fused to the C-terminus of Psb35 (hereafter Psb35-CLFP). We initially analyzed the membranes of the strain using 2D CN/SDS-PAGE to assess whether this fusion protein accumulates and is present in similar complexes like the original Psb35. The scanning of the native gel using the confocal microscope equipped with a special objective documented the presence of bands containing CLFP and subsequent 2D blotting confirmed the association of Psb35-CLFP with CP47m, RC47 and PSII(1) (Supplementary Fig. S5). Thus, we subjected the exponentially grown autotrophic cells of the mutant to confocal microscopy (Fig. 4) and successfully detected the signal of Psb35-CLFP. Although distribution of the labeled protein was mostly uniform, there were some discrete spots with higher CLFP and lower Chl fluorescence intensity (Fig. 4, white arrows).
Cellular localization of Psb35 fluorescently labeled using CLFP in WT and ΔCP47 background strains. The autofluorescence Chl signal (upper panels), the signal of Psb35-CLFP (middle panels) and their overlap (lower panels) are shown for WT (the first panels from left), Psb35-CLFP (the second panels from left), ΔCP47 (the third panels from left) and ΔCP47/Psb35-CLFP (the fourth panels from left) strains. White arrows indicate spots in the signal of Psb35-CLFP, which does not overlap with autofluorescence. The scale bars represent 2 µm.
The average CLFP fluorescence from Psb35-CLFP cells was rather low and the reason for it could be related to a generally low fluorescence of the protein, to its low abundance or to an energy transfer from CLFP to nearby pigments, for instance phycobilisomes (PBS). To evaluate these possibilities, we also constructed a strain expressing Psb35-CLFP additionally lacking CP47, which contains much lower content of fluorescing PSII-bound Chls since it lacks assembled PSII and accumulates the unassembled CP43 (Komenda et al. 2004). The CL-fluorescent regions, not overlapping with the Chl fluorescence, were more extensive than in the WT background strain (Fig. 4). These data suggest that the Psb35-CLFP mostly co-localizes with PSII complexes, but a fraction of the protein occurs in the regions with missing PSII, which are more apparent in the CP47-less strain. Moreover, the CLFP signal in the double mutant was about twice as intense as that in the single CLFP mutant (Supplementary Fig. S6A) despite similar levels of the expressed Psb35-CLFP (Supplementary Fig. S6B). This suggests a CP47-dependent fluorescence quenching of CLFP fluorescence, which could be related to an energy transfer to other pigments like membrane-bound PBS.
PSII function and biogenesis in the Psb35-less mutant
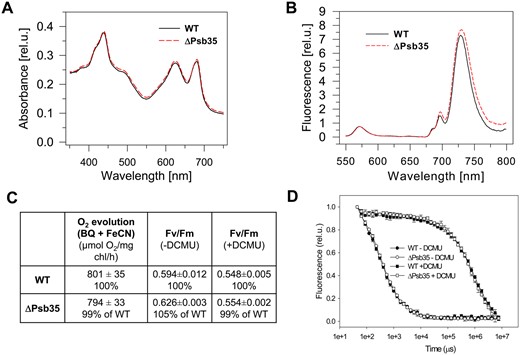
To gain insight into the function of the protein, we constructed a Synechocystis strain in which the psb35 gene was inactivated by a gentamicin-resistant cassette. The fully segregated Psb35-less strain (ΔPsb35 strain, for PCR confirmation see Supplementary Fig. S7) was characterized by several methods. Comparison of the absorption spectra (Fig. 5A) and 77K Chl fluorescence spectra (Fig. 5B) between WT and ΔPsb35 showed no significant differences in pigmentation and PSII/PSI content. The strain also exhibited the same PSII oxygen-evolving activity, PSII quantum yield FV/FM (Fig. 5C) and the variable fluorescence decay (Fig. 5D) documenting no difference in the function of PSII between WT and ΔPsb35. We also compared the pattern of membrane complexes in both strains using 2D CN/SDS-PAGE (Fig. 6). In WT membranes, we observed a standard set of pigment-binding complexes, namely (in the order of the decreasing size) RCCS, PSI(3), PSII(2), PSI(1), PSII(2), RC47, CP47m′ (Fig. 6)(Fig. 2, black arrow), CP47m and CP43m. This set was quite similar for the ΔPsb35 strain but the CP47m′ was missing, which confirmed its identity as the CP47m containing Psb35. In addition, the mutant contained more RCII complexes lacking both CP47 and CP43, which are typical by the presence of D1 intermediate (iD1, Fig. 6, white arrows). This suggests some limitation in the availability of CP47 for the continuation of PSII assembly beyond RCII.
Whole cell absorption spectra (A), 77K Chl fluorescence spectra (B), PSII-mediated oxygen evolution and quantum yield of PSII photochemistry Fv/Fm (C) and variable fluorescence decay in the absence and presence of PSII inhibitor (D) of photoautotrophic WT and ΔPsb35 cells cultivated for 3 d under 30 µmol photons m−2 s−1. (A) Whole cell absorption spectra of WT (black solid line) and ΔPsb35 (red dashed line) liquid cultures are shown after normalization to the optical densities at 750 nm. (B) For 77K fluorescence spectra equal amounts of cells from WT (black solid line) and ΔPsb35 (red dashed line) liquid cultures were frozen in liquid nitrogen and excited at 435 nm. Spectra were normalized to the emission peak of the internal standard rhodamine at 570 nm. (C) The light-saturated rate of oxygen evolution measured in the presence of 2.5 mM p-benzoquinone and 2 mM potassium ferricyanate was measured in the exponentially grown cultures using Clark electrode, and values represent means of three biological replicates and three measurement each ± SD; Fv/Fm values in the exponentially grown cultures were obtained using P.S.I. modulated fluorimeter, and values represent means of three biological replicates ± SD. (D) Variable fluorescence decay curves in the exponential grown cultures of WT (closed symbols) and ΔPsb35 (open symbols) either untreated (circles) or treated (squares) with DCMU were measured using P.S.I. modulated fluorimeter; values represent means of three biological replicates ± SD.
To evaluate the effect of ΔPsb35 mutation on the synthesis of membrane proteins, especially those of PSII, the WT and mutant cells were radioactively labeled in vivo with a mixture of [35S] Met/Cys. The membranes isolated from these cells were analyzed by 2D CN/SDS-PAGE, and the gel was stained by Coomassie Brilliant Blue (2D CBB stain), dried and subjected to autoradiography (Supplementary Fig. S8). The pattern of labeled proteins in autoradiograms from both strains was generally similar; nevertheless, there were some subtle but reproducible differences between them. First, the mutant exhibited higher level of labeling of unassembled forms of the D1 protein (Supplementary Fig. S8, arrow triplet, for identification see Kiss et al. 2019). Second, there was a different pattern of labeled CP47 in the WT and mutant (Supplementary Fig. S8, numbered white arrows): both strains contained equally labeled CP47 bands in the RCCS (arrows 1 and 8), PSII(2) (arrows 3 and 9), PSII(1) (arrows 4 and 10) and CP47m (arrows 7 and 12); in comparison with the mutant WT contained more intensive labeled bands of CP47 within PSI(3)-CP47m′ (arrow 2) and within two larger CP47ms (arrows 5 and 6), which represent most probably CP47m′ and CP47m′ associated with Hlips (black arrow 16). Moreover, the mutant CP47m, corresponding to the labeled CP47 (arrow 11), had the higher mobility than the CP47m′-Hlips complex in WT, as it most probably contained just HliA/B but not Psb35. Third, the mutant showed higher intensity of labeled HliA/B within PSII(1) (black arrow 15) than WT (black arrows 13). Finally, the band belonging to the newly synthesized PSI proteins PsaA/B was more intensively labeled in WT (black arrow 17) than in the mutant (black arrow 18). The described differences indicated that Psb35 modifies the overall pattern of synthesis/degradation of Chl-binding proteins, namely PsaA/B, D1, CP47 and HliA/B.
Effect of Psb35 deletion on the Hlip and CP47 stability
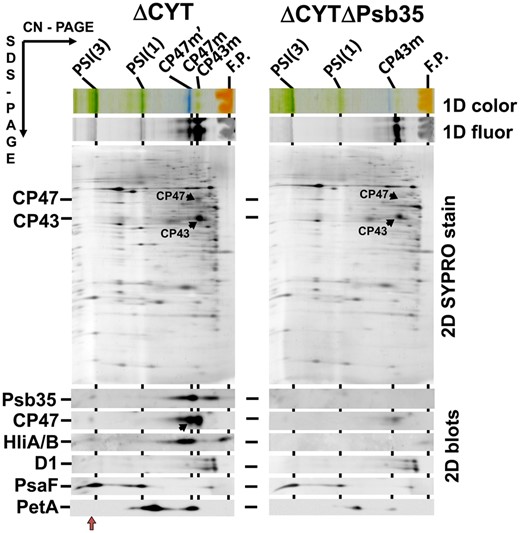
To explore an effect of Psb35 on the accumulation of CP47m and Hlips in more detail, we used the previously characterized strain lacking the psbEFLJ operon, which encodes several small PSII proteins, including cytochrome b-559 subunits (ΔCYT; Komenda et al. 2004). This strain cannot accumulate the D2 protein and therefore cannot form any PSII assembly intermediate complexes. Unlike the single mutant, the double ΔCYT/ΔPsb35 mutant grew slower under standard growth light conditions in the presence of glucose, accumulated much less PBS (Supplementary Fig. S9) and contained much less unassembled CP47, CP43, HliA/B, PSI trimer and also cytochrome f than the single ΔCYT mutant (Fig. 7). These data surprisingly suggested that not only Chl but also heme accumulation was impaired after inactivation of Psb35.
2D analysis of membrane proteins of WT and ΔPsb35. Membranes were analyzed by 2D CN/SDS-PAGE. After the first dimension, the gel was photographed (1D color) and scanned for Chl fluorescence (1D fluor). After the separation in the second dimension, the 2D gel was stained using Sypro Orange (2D SYPRO stain) and blotted to a poly(vinylidene fluoride) membrane (2D blots) and Psb35, CP47 and D1 were detected by the specific antibodies. Designation of complexes as in Figs. 1, 2; the black arrow designates CP47 within the CP47m′ with bound Psb35, and the white arrows designate iD1 in the RCII complexes lacking both CP47 and CP43. Each loaded sample contained 5 µg of Chl.
2D analysis of membrane proteins of ΔCYT and ΔCYTΔPsb35 strains. Membranes were analyzed by 2D CN/SDS-PAGE. After the first dimension the gel was photographed (1D color) and scanned for Chl fluorescence (1D fluor). After the separation in the second dimension, the 2D gel was stained using Sypro Orange (2D SYPRO stain) and blotted to a poly(vinylidene fluoride) membrane (2D blots) and Psb35, CP47, HiA/B, D1, PsaF and PetA were detected by the specific antibodies. Designation of complexes as in Figs. 1, 2; the black arrow in the blot designates CP47 within the CP47m′, the red arrow PSI(3)-CP47m′ complex. Loading of the samples was based on the same OD750 nm corresponding to 3 μg of Chl of ΔCYT.
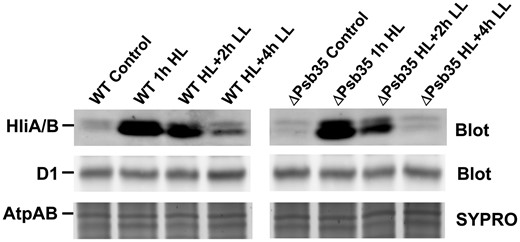
The stabilization effect of Psb35 on the unassembled CP47 and Hlips was also apparent in a previously constructed strain lacking CP43 (encoded by the psbC gene), which accumulates the PSII assembly complex RC47, CP47m and Hlips even without exposure to stress conditions (Boehm et al. 2012). We additionally inactivated the psb35 gene in this strain, and then, we compared the content of CP47 and HlipA/B in the single and double mutants. The analysis of membranes from both strains using 2D CN/SDS-PAGE (Supplementary Fig. S10) showed that the amount of HliA/B bound to both RC47 and CP47 was remarkably lower in the double mutant than in the CP43-less single mutant. In addition, the amount of CP47m was decreased in the absence of Psb35. To demonstrate the stabilization effect of Psb35 on the HliA/B proteins in WT strain, we exposed the WT and ΔPsb35 strains to higher irradiance (300 µmol photons m−2 s−1) and after 1 h of illumination we transferred the cells back to growth irradiance (40 µmol photons m−2 s−1). Monitoring of the HliA/B content during the experiment documented the faster disappearance of Hlips in the mutant than in WT after decreasing the light intensity (Fig. 8).
1D analysis of membrane proteins of WT and ΔPsb35 strains treated with high light (300 µmol photons m−2 s−1) for 1 h and then transferred back to growth irradiance for 2 and 4 h. Membranes were analyzed by 1D SDS-PAGE, stained by SYPRO Orange (SYPRO) and blotted to a poly(vinylidene fluoride) membrane (Blot) and D1 and HliA/B were detected by the specific antibodies. Staining of AtpA/B proteins (SYPRO) is shown to document the equal loading. Two micrograms of Chl were loaded for each strain.
Accelerated pigment bleaching in the mutant cells incubated in the dark
In summary, the present results provided the strong experimental evidence that removal of Psb35 leads to a destabilization of CP47m and HliA/B; therefore, we expected a retardation of the mutant growth in comparison with WT under increased irradiance, when HliA/B accumulates. However, the growth of cells under both growth (40 µmol photons m−2 s−1) and high (300 µmol photons m−2 s−1) irradiance was very similar in both strains (Fig. 9A). Moreover, the growth fitness of the ΔPsb35 strain did not differ from WT under a variety of other growth conditions, including intermittent light, low or high temperature. However, we found that after 10 d of dark incubation in the absence of any source of reduced carbon, the level of Chl and carotenoids was substantially lower in the mutant than in WT as judged from whole cell spectra (Fig. 9B). 2D gel showed a larger depletion of PSI(3) and PSII(1) in the mutant when compared with WT (Fig. 9C, red arrows) and, interestingly, the most abundant PSII complex in the cells of both strains became RC47 while CP43 was depleted, especially in the mutant. Thus, in the dark, the absence of Psb35 promoted the loss of both PSII and PSI Chl-binding complexes.
Growth comparison (A), absorption spectra of WT and ΔPsb35 cells before and after incubation in the dark for 10 d (B) and 2D analysis of their membrane proteins (C). (A) Drops of WT and ΔPsb35 cultures at the indicated OD750 nm were pipetted on a solid agar plate and the plate was photographed after 3 d of cultivation. (B) Cells of both strains before and after 10 d incubation in the dark were measured using Shimadzu UV-3000 based on the same OD750 nm. The spectra of dark incubated cells were shifted by 0.05 for better illustration of the changes. (C) Membranes of both strains after the dark incubation were analyzed by 2D CN/SDS-PAGE. After the first dimension the gel was photographed (1D color) and scanned for Chl fluorescence (1D fluor). Bands of PSI(3) and PSII(1) are designated with red arrows to emphasize the differences in their intensities between the strains. After the separation in the second dimension the 2D gel was stained using Sypro Orange (2D SYPRO stain), electroblotted and CP47 and CP43 detected using specific antibodies. Four micrograms of Chl were loaded for each strain.
Discussion
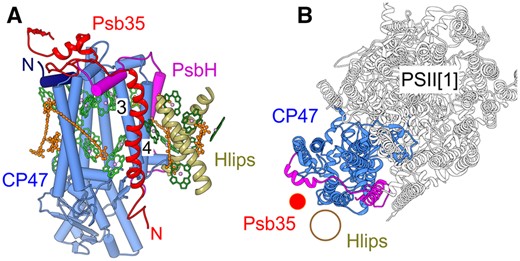
We have discovered a new protein, designated here as Psb35, that binds to CP47 during assembly of PSII. Psb35 is a small 11-kDa protein, which is predicted to contain a single transmembrane helix with its N-terminus exposed to the lumenal side of the membrane. The binding of Psb35 to CP47 is not dependent on PsbH, while it has the protective effect on HliA/B proteins, which bind to CP47 in a PsbH-dependent manner (Fig. 1; Promnares et al. 2006). A small amount of protein can also be detected in PSII dimers (Fig. 2B). Taking into account the structure of dimeric PSII (Umena et al. 2011), these data suggest a possible location for Psb35 in the vicinity of the 3rd and 4th transmembrane helix of CP47 (Fig. 10). The protein has a 60-amino-acid residue long C-terminus exposed to cytoplasm, which could also play a functional role. Modeling of the protein using Phyre did not reveal any typical structural/functional domain at the C-terminus. Nevertheless, Psb35 was found to stabilize unassembled CP47 and HliA/B proteins and a possible explanation for this effect is that long C-terminus hides the N-terminal parts of HliA/B and CP47 from the premature proteolytic attack. Indeed, the best characterized cyanobacterial membrane proteolytic FtsH2/FtsH3 complex, which also regulates the level of unassembled CP47 (Komenda et al. 2006), has been shown to degrade at least the D1 protein processively from the cytoplasmatically exposed N-terminus (Komenda et al. 2007). It is probable that this mechanism of degradation is also valid for CP47 and HliA/B and therefore hiding their N-termini by the long C-terminus explains well the protective effect of Psb35 on both proteins. The protective effect of Psb35 is best discernible in mutants which accumulate CP47m, i.e. PsbEFLJ- and CP43-less strains. Given this proposed function of the protein, it is conceivable that fusion of the N-terminus of an Hlip with Psb35 identified in two thermophilic cyanobacterial strains (Synechococcus OS-A′ and OS-B′; Kilian et al. 2008) might represent even a more efficient way of Hlip protection. Actually, Hlips seem to possess a tendency to fusion as the Hlip domain is also present in ferrochelatase enzyme that catalyzes the insertion of iron into protoporphyrin IX to produce heme. Protoporphyrin IX is the last common biosynthetic precursor of both heme and Chl and recent work has pointed toward a potential role of the Chl-binding Hlip domain in the regulation of ferrochelatase (Pazderník et al. 2019).
Model of the Psb35 localization within CP47m (A) and PSII monomer (B). The model is based on the PSII crystal structure (PDB 3WU2; Umena et al. 2011) and putative location of Hlips based on CP29 binding to CP47 within the plant PSII-LHCII supercomplex (PDB 5XNL; Su et al. 2017). (A) A view on CP47 parallel with the membrane plane; CP47 in light blue, its N-terminus in dark blue; PsbH in magenta, Psb35 in red and Hlips in khaki, Chls in green and β-carotene molecules in orange; numbers designate the helices 3 and 4 of CP47. (B) A view on PSII monomer perpendicular to the membrane plane, color code as in (A); for simplification, helices of Psb35 and Hlips are designated just by red and khaki circles, respectively.
Our previous experiments showed that the accumulation of PSI trimers and CP47 is dependent on the availability of de novo synthesized Chl (Sobotka et al. 2008, Hollingshead et al. 2016). Perhaps it is not a coincidence that Psb35 was also identified as a component of a novel supercomplex consisting of both these Chl-binding complexes. Although we cannot completely exclude that this complex is artifactual, we consider it improbable since it was isolated using His- and FLAG-specific affinity chromatography (Fig. 1B and Supplementary Fig. S3) and it was detected in the native membranes of WT using radioactive labeling (Supplementary Fig. S8, arrow 2) and PsbEFLJ- and CP43-less strains using immunodetection (Fig. 7 and Supplementary Fig. S10, red arrows). Moreover, CP47m′ is not simply attached to the trimer as it would be expected from unspecific interaction, but it is partly inserted between two monomers of the trimer. The physiological significance of the PSI(3)-CP47m′ supercomplex is presently unclear. Although Psb35 was detected in the complex, its formation is not dependent on Psb35 as we isolated a similar complex from the His-CP47 expressing strain lacking both CP43 and Psb35. This supercomplex may reflect an earlier transient assembly intermediate during the formation of the PSII complexes as we have previously characterized a putative later assembly intermediates consisting of PSI(3) and two monomeric PSII core complexes from high-light exposed cells expressing FLAG-tagged auxiliary factor Psb28 (Bečková et al. 2017a). As we proposed before, the robust photochemistry of PSI(3) could provide a safe escape for the energy absorbed by the unassembled CP47, thus protecting the antenna from oxidative damage until it is properly associated with other proteins to form the functional PSII.
Alternatively, the PSI(3)-CP47m′ supercomplex might also represent an assembly intermediate for the formation of PSI(3). From this point of view, it is interesting that the detailed structure of the PSI trimer from Synechocystis revealed its unexpected asymmetry, which could reflect a separate formation of the tighter dimer and a subsequent attachment of a monomer, which remains bound more loosely within the trimer than remaining two monomers (Malavath et al. 2018). And just this loosely bound monomer could be the one that is in contact with CP47 in the observed PSI(3)-CP47m′ supercomplex. So, the formation of the PSI(3) could be facilitated by the attachment of CP47m. Indeed, the CP47-less strain is deficient in PSI (Bečková et al. 2017a) and namely in the trimer [see comparison of PSI(3) in WT and ΔCP47 membranes in Fig. 2].
In fact, the close interaction between CP47 and PSI may explain the depletion of PSI (and CP43) in the dark. Under these conditions, the biosynthesis of new Chl molecules is quite limited and remaining ones are mostly consumed by the newly synthesized CP47 while their availability for other PSII Chl-binding proteins is minimized. When CP47 cannot be assembled, it is quickly degraded even in the dark (Komenda et al. 2006) and during this degradation, a part of the released Chls becomes degraded. This missing Chl required for CP47 synthesis can be compensated by its withdrawal from the neighbor PSI(3), which is afterward converted into monomers and finally degraded like observed in the absence of phosphatidylglycerol (Kopečná et al. 2015). Also, the fast turning-over D1 protein, which most probably uses Chls released from the degraded old D1 copy, may obtain the lost Chl molecules from other Chl-binding proteins like PSI. Thus, finally the RC47 complex is maintained while PSI is depleted. Depletion of CP43 under these conditions should also be expected as there is a relationship between biogenesis of CP43m and PSI monomer (Komenda et al. 2012a, Kopečná et al. 2015). Obviously, when CP47m and associated Hlips are stabilized by Psb35, the loss of Chl and its withdrawal from PSI is partially suppressed. As a result, a slower Chl bleaching is observed in the dark in the WT cells in comparison with the Psb35 cells.
The longer-living CP47m with bound Psb35 could also stabilize some components of Chl-biosynthesis pathway as suggested by Chl deficiency of the CP47 strain (Bečková et al. 2017a), and this may prolong the dark synthesis of Chl and retard its disappearance. From this point of view, it is interesting that the destabilization of CP47 in the strain ΔCYT lacking the psbEFLJ operon due to the Psb35 elimination seems to have additional effect on the accumulation of both Chl and heme. ΔCYT cannot assemble PSII and accumulates a small amount of unassembled CP47, but the additional removal of Psb35 led to the nearly complete disappearance of CP47 but also to depletion of CP43, PSI and even PBS and cytochrome b6f complex (Fig. 7 and Supplementary Fig. S9). Only accumulation of the unassembled D1 protein was affected much less confirming the idea that synthesis of D1 is not dependent on the newly synthesized Chl (Hollingshead et al. 2016). The reason for such a strong phenotype in the double mutant is not clear. We speculate that CP47 and cytochrome b-559 could represent primary acceptors of both major products of the pathway (i.e. Chl and heme) and in their absence these products cause inhibition of the whole tetrapyrrole biosynthetic pathway, for instance via a feedback regulation. In accordance with this, the HPLC analysis confirmed a similar 30% depletion of heme and coproporphyrinogen III while the accumulation of Mg protoporphyrin IX methylester and depletion of the monovinyl- and divinyl-protochlorophyllide (Supplementary Fig. S9B) pointed to the inhibition of the Chl cyclase reaction previously seen in the Psb28-less mutant (Dobáková et al. 2009).
The finding that Psb35 contributes to the stability of the cyanobacterial Chl-binding proteins in the dark might explain why in many oceanic cyanobacterial species genes encoding Psb35 homologues are missing. These species are exposed to the regular day–light cycles and should never experience long-term dark periods. By contrast, fresh water and terrestrial cyanobacteria may often be exposed to such dark periods. Indeed, the cyanobacterium Gloeobacter violaceus, which belongs to rock-dwelling organisms and is optimized for growth in low light (Mareš et al. 2013), possesses even several genes for Psb35 homologs.
Psb35 homologs are also missing in algae and higher plants. A possible explanation could be the absence of Hlip-like proteins associated with CP47 in these organisms. While most cyanobacteria contain at least four members of the Hlip family and those homologous to HliA/B are expected to bind to CP47, the Arabidopsis genome seems to contain just two genes encoding single helix Hlip-like OHP proteins (Hey and Grimm 2020), which are similar to HliC/HliD pair associated with Chl synthase and D1–D2 containing RCII complex (Komenda and Sobotka 2016). While it lacks homologs of HliA/B, it contains genes encoding other plant LHC-like proteins like two-helix LILs and three-helix ELIPs (Jansson 1999), which have been found to associate with Chl-biosynthesis enzymes (Tanaka et al. 2010, Hey et al. 2017, Zhou et al. 2017). Existing data on the function and location of LILs and ELIPs (Rochaix and Bassi 2019) do not exclude that these proteins might also associate with CP47 similarly to HliA/B in cyanobacteria but they already contain the additional helix not binding pigments that may play an analogous stabilizing role as the cyanobacterial Psb35. Although the second helixes of various LHC-like proteins are largely divergent (Engelken et al. 2010), the comparison of amino acid sequences of Psb35 and the second helix of SEP1 from Arabidopsis (At4g34190) and other higher plants showed an apparent similarity indicating their possible evolutionary relationship (Supplementary Fig. S11).
Concerning the localization of Psb35 within the Synechocystis cell, using the fluorescently labeled Psb35-CLFP, we showed its co-location with red wavelengths emitting PSII. However, there were few spots at the cellular periphery exhibiting dominant green fluorescence. Size of these spots increased when a large portion of the PSII-related Chl fluorescence was eliminated by deletion of the gene for CP47. The presence of Psb35 in PSII complexes containing assembly factors, including Hlips, clearly speaks for its involvement in the biogenesis of PSII. Interestingly, cells of G. violaceus devoid of TMs have their photosynthetic complexes segregated into ‘green domains’ meanwhile the other proteins and their complexes, including photosystem assembly factors, have been detected in ‘orange domains’ of the plasma membrane (Rexroth et al. 2011). The peripheral spots enriched in Psb35 could represent a Synechocystis analogy of such orange biogenesis domains.
Materials and Methods
Cyanobacterial strains and construction of mutants
In this work, we used the Synechocystis GT-P sub-strain (Tichý et al. 2016) as WT strain and other previously described strains listed in Supplementary Table S1. The ssl2148-less strain ΔPsb35 was constructed by replacing the psb35 (ssl2148) gene by a gentamicin antibiotic resistance cassette. The sequences upstream and downstream (500 bp) of the psb35 gene were amplified using two sets of primers (Supplementary Table S3), and fusion PCR in conjunction with megaprimers (Ke and Madison 1997) was used to anneal these on either side of the gentamicin-resistant cassette. The resulting PCR product (Supplementary Fig. S7) was used for the transformation of WT and the transformed cells were fully segregated on BG11 plates with increasing concentrations of gentamicin. The strain expressing Psb35 fused with 3xFLAG at the C-terminus under psbAII promoter was constructed using pPD-CFLAG vector described in Chidgey et al. (2014) and primers shown in Supplementary Table S3. The ΔPsb35 strain was then transformed with the prepared pPD-psb35-CFLAG construct, and the genetic locus was segregated on plates with kanamycin. To construct a mutant expressing the mClover3 fluorescence protein (Bajar et al. 2016) fused to the C-terminus of Psb35 via a short SAGGSGGSV spacer (Supplementary Fig. S12A), we transformed WT or CP47-less strains with the commercially synthesized psb35-mClover3-Ery construct cloned into the pUC57-mini vector (Genscript, USA; Supplementary Fig. S12B). The plasmid contained the particular sequences of the fused genes with the short spacer followed by the erythromycin-resistant cassette, and this all flanked from both sides by 500 bp upstream and downstream regions of the psb35 gene. Strains were segregated on plates containing erythromycin. To construct multiple mutants combining the psb35 deletion or FLAG tagging with other mutations, the existing mutants (see Supplementary Table S1) were transformed using chromosomal DNA, PCR product or commercial plasmid containing particular psb35 version and the mutants were segregated on plates containing particular antibiotics (Komenda et al. 2008). The correct sequence of the construct was confirmed by DNA sequencing in all new transformants.
Cultivation conditions of cyanobacterial strains
The strains were grown in 100 ml of liquid BG11 medium using 250-ml conical flasks in a rotary shaker under 40 µmol photons m−2 s−1 (‘normal light’) at 29°C in liquid BG11 medium. Non-autotrophically cultured PSII- or PSI-less mutants were supplemented with 5 mM glucose. For protein purification, 2 or 4 l of cells were grown photomixotrophically in 10-l flasks under 100 µmol of photons m−2 s−1 in BG11 medium supplemented with 5 mM glucose. The cell culture was agitated with magnetic stirrer and bubbled with air.
For the dark and high light treatment, the cells were first cultivated under normal light conditions until they reached exponential phase [0.6–0.8 optical density (OD) at 750 nm]. At that point, the cultures were transferred either into an incubator, in which the culture was stirred with magnet stirrers in the dark (dark incubation), or into a shaker under 300 μmol photons m−2 s−1 (‘high light’).
Growth assays on agar plates
Growth assays were performed as described in Bečková et al. (2017a). Briefly, after reaching exponential phase, the cultures were diluted to OD750 nm = 0.1, 0.05 and 0.025 and spotted on autotrophic BG11 agar plates containing 10 mM TES buffer. The plates were incubated at 29°C and either at 40 µmol photons m−2 s−1 or at 300 µmol photons m−2 s−1 (high light) for 3 d. We also tested other conditions like combination of high light and low temperature (17°C) or intermittent light conditions (5 min dark and 5 min 300 µmol photons m−2 s−1).
Whole cell absorption spectroscopy and pigment determination
Absorption spectra of whole cells were measured at room temperature using a UV-3000 spectrophotometer (Shimadzu, Japan) and were measured in the cultures with the identical OD750 nm. For routine Chl determination, pigments were extracted from cell pellets with 100% methanol and the Chl concentration was determined spectroscopically (Wellburn 1994). Detailed analysis of pigments, heme and Chl-biosynthesis precursors using HPLC was performed as described in Pilný et al. (2015) and Kiss et al. (2019).
Low temperature fluorescence spectroscopy
Chl fluorescence emission spectra 77K were measured using an Aminco Bowman Series 2 luminescence spectrometer (Spectronic Unicam, Rochester, NY, USA) and fluorescence spectra were recorded in the range 600–800 nm. The spectra were measured in cultures with the identical OD750 nm and normalized using 1 µM rhodamine as an internal standard.
PSII activity
The activity of PSII was measured by assessing the oxygen evolution rate using a Clark-type electrode in the presence of the artificial electron acceptors p-benzoquinone (2.5 mM) and potassium ferricyanide (1 mM) (Sigma, USA). The data are averages ± SD of three biological experiments, three repetitions in each.
Electron transport kinetics within PSII complexes were followed by the emission and subsequent relaxation kinetics of flash induced Chl fluorescence in the absence and presence of 10 µM DCMU as described in Vass et al. (1999) using P.S.I. modulation fluorimeter (P.S.I., Czech Republic). The FV/FM values were determined by the same method.
Transmission electron microscopy
Freshly prepared complexes were used for electron microscopy. The specimen was placed on glow-discharged carbon-coated copper grids and negatively stained with 2% uranyl acetate, visualized by JEOL JEM-2100F transmission electron microscope (JEOL Japan, using 200 kV at 20,000× magnification) and processed by image analysis. TEM images were recorded by a bottom-mount Gatan CCD Orius SC1000 camera, corresponding to a pixel size of 3.4 Å. Image analysis was carried out using RELION (Scheres 2012). The selected projections were rotationally and translationally aligned and treated by empirical Bayesian approach in combination with classification procedure to refine 2D class averages. The projection was overlaid with a cyanobacterial X-ray models of the PSI (1JB0, Jordan et al. 2001 and CP47 Umena et al. 2011).
Confocal imaging
Images were acquired using Laser Scanning Microscope LSM 880 (Zeiss, Germany) using a Plan-Apochromat 63×/1.4 Oil DIC M27 objective and EC Plan-Neofluar 5×/0.16, for cell culture and gel strips, respectively. Live cell images were acquired in two scans. Emissions of Chl (695–758 nm) and CLFP (499–526 nm) were simultaneously acquired using excitation at 488 nm (MBS 488) while emission of PBS (642–677) was acquired using excitation at 633 nm. Image settings were: 512 × 512 pix, 8-bit, pix dwell = 33 µs, pinhole = 50 µm (e.g. 488 nm), 63 µm (e.g. 633 nm).
Gel strip images were acquired in two tile scans, excited either at 488 nm for the detection of CLFP (emission range 499–526 nm) and Chl ( emission range 624–695 or 695–758 nm), or at 633 nm for the detection of PBS (emission range 642–677 nm). Image settings was: 1,918 × 94 pix, 8-bit, pix dwell = 11 µs, pinhole = 601 µm. Cell and gel strip images were processed using ImageJ, and gel images were then aligned with the camera images.
Radioactive labeling
Radioactive pulse labeling of the cells was performed at 500 µmol photons m−2 s−1 and 30°C using a mixture of [35S]Met and [35S]Cys (Hartmann Analytic Gmbh, Germany) as described previously (Dobáková et al. 2009).
Preparation of membranes and trypsin treatment
Small-scale membrane fractions were prepared by breaking the cells with zirkonia/silica beads using a Mini-Beadbeater (BioSpec, USA) according to the procedure described in Komenda and Barber (1995), but the cells were resuspended and broken in 25 mM MES/NaOH, pH 6.5, 10 mM CaCl2, 10 mM MgCl2, 25% glycerol (buffer B). The trypsinization of isolated membranes was performed as described in Dobáková et al. (2009).
FLAG- and His-tag protein purifications
Large-scale membrane preparations for the purification of proteins in the buffer B containing 0.04% n-dodecyl-β-d-maltoside (β-DDM) and a protease inhibitor cocktail (Roche, USA, or Sigma-Aldrich, USA) were isolated using Mini-Beadbeater-16 (BioSpec, USA) as described in Koskela et al. (2020). The membranes were solubilized with β-DDM and FLAG-tagged proteins were isolated using the anti-FLAG M2 affinity gel (Sigma-Aldrich, USA) as described in detail in Koskela et al. (2020). His-tagged CP47m, CP47m/ΔPsbH and CP43m were isolated using a single-step Ni-affinity purification from strains expressing His-tagged PsbH, CP47 or CP43 but unable to assemble PSII (for more precise genetic defining the strains see Supplementary Table S1). The β-DDM-solubilized membranes were loaded on the Ni-affinity resin (Protino Ni–NTA Agarose, Macherey-Nagel, Germany), and after an intensive washing with buffer B containing 0.04% β-DDM and 30 mM imidazole, the proteins were eluted with buffer B containing 0.04% β-DDM and 150 mM imidazole.
Analysis of proteins and their complexes
The composition of membrane protein complexes was analyzed as described in Komenda et al. (2012b) by clear native (CN) electrophoresis in a 4–14% gradient polyacrylamide gel in combination with SDS-PAGE in a denaturing 12–20% gradient gel containing 7 M urea. One-dimensional SDS-PAGE was carried out in the same 12–20% denaturing gel. The separated proteins were visualized by staining with either CBB or SYPRO Orange and detected by protein mass spectrometry (MS) or immunoblotting. The primary antibodies against D1, D2, CP47, CP43, PsbE and PsbF used in this study and prepared in rabbits were previously described by Komenda et al. (2008). Furthermore, we employed the antibody against Synechocystis Ycf48 (Yu et al. 2018) and PsbH (Bumba et al. 2005), Synechocystis PCC 6301 PsaD (Xu et al. 1994) and commercial antibodies anti-PetA, anti-HliA (both Agrisera, Sweden), anti-FLAG (Abgent, USA) and anti-GFP (Abcam, UK) also recognizing CLFP. We also prepared the antibody against PsaF specific to peptide 50–61 of the Synechocystis protein and against Psb35 specific to the C-terminal region of the Synechocystis protein. The blots were developed using the anti-rabbit secondary antibody conjugated with horse radish peroxidase (Sigma-Aldrich, USA) and chemiluminescence substrate Immobilon Crescendo (Merck, USA).
Protein identification by mass spectrometry
The MS analyses of protein bands excised from the gel were done on a NanoAcquity UPLC (Waters, USA) online coupled to an ESI Q-ToF Premier mass spectrometer (Waters, USA) as described in Janouškovec et al. (2013).
Supplementary Data
Supplementary data are available at PCP online.
Funding
Czech Science Foundation (Project No. 19-29225X); European Research Council project Photoredesign (No. 854126); and European Regional Development Fund (Project No. CZ.02.1.01/0.0/0.0/15_003/0000441 to Z.G.).
Acknowledgments
G.P.A. and J.K. designed the research. G.K., E.K., Z.G., M.B., J.Kno., L.B., R.S. and J.K. performed the experiments. G.P.A., G.K., Z.G., R.K., R.S. and J.K. analyzed the data. J.Kno., G.P.A., R.S. and J.K. wrote the article. We are grateful to J. Pilný for the analysis of heme and Chl precursors and P. Nixon for a critical reading of the manuscript.
Disclosures
The authors have no conflicts of interest to declare.