-
PDF
- Split View
-
Views
-
Cite
Cite
Brigitta M Kállai, Hana Kourová, Jana Chumová, Csaba Papdi, Lucie Trögelová, Olga Kofroňová, Pavel Hozák, Vlada Filimonenko, Tamas Mészáros, Zoltan Magyar, Laszlo Bögre, Pavla Binarová, γ-Tubulin interacts with E2F transcription factors to regulate proliferation and endocycling in Arabidopsis, Journal of Experimental Botany, Volume 71, Issue 4, 7 February 2020, Pages 1265–1277, https://doi.org/10.1093/jxb/erz498
Close - Share Icon Share
Abstract
γ-Tubulin is associated with microtubule nucleation, but evidence is accumulating in eukaryotes that it also functions in nuclear processes and in cell division control independently of its canonical role. We found that in Arabidopsis thaliana, γ-tubulin interacts specifically with E2FA, E2FB, and E2FC transcription factors both in vitro and in vivo. The interaction of γ-tubulin with the E2Fs is not reduced in the presence of their dimerization partners (DPs) and, in agreement, we found that γ-tubulin interaction with E2Fs does not require the dimerization domain. γ-Tubulin associates with the promoters of E2F-regulated cell cycle genes in an E2F-dependent manner, probably in complex with the E2F–DP heterodimer. The up-regulation of E2F target genes PCNA, ORC2, CDKB1;1, and CCS52A under γ-tubulin silencing suggests a repressive function for γ-tubulin at G1/S and G2/M transitions, and the endocycle, which is consistent with an excess of cell division in some cells and enhanced endoreduplication in others in the shoot and young leaves of γ-tubulin RNAi plants. Altogether, our data show ternary interaction of γ-tubulin with the E2F–DP heterodimer and suggest a repressive role for γ-tubulin with E2Fs in controlling mitotic activity and endoreduplication during plant development.
Introduction
γ-Tubulin is a highly conserved microtubule nucleator in eukaryotic cells. The microtubule-independent role of γ-tubulin in cell cycle regulation and in nuclear processes shows γ-tubulin to be a multifunctional protein (Oakley et al., 2015). The presence of γ-tubulin in nuclei of plant (Binarova et al., 2000) and animal cells (Horejsi et al., 2012) suggested its nuclear functions. γ-Tubulin interacts with the DNA repair protein Rad51 (Lesca et al., 2005) and with the tumour suppressor protein C53 (Horejsi et al., 2012), and forms a well-characterized complex with BRCA1 (Gomez and Hergovich, 2014). Evidence was provided for the role of γ-tubulin in chromatin remodelling (Vazquez et al., 2008) and in transcription (Hoog et al., 2011). Functions of γ-tubulin in organization of the nuclei through interaction with lamins (Rossello et al., 2016) and SUN proteins (Chumova et al., 2019) are suggested. Defective cell cycle progression and coordination of mitotic events were observed in γ-tubulin mutants of Schizosaccharomyces pombe (Hendrickson et al., 2001) and in Aspergillus (Prigozhina et al., 2004). Aspergillus γ-tubulin mutants showed impaired inactivation of the anaphase-promoting complex (APC) and coordination of mitosis with the spindle assembly checkpoint (Edgerton-Morgan and Oakley, 2012; Edgerton et al., 2015). In acentrosomal plant cells, γ-tubulin is broadly distributed in the cytoplasm with microtubules and membranes and has a role in microtubule nucleation from dispersed sites (Binarova et al., 2006; Pastuglia et al., 2006). Besides its microtubule nucleation function, microtubule-independent roles of γ-tubulin in plant cell division have also been suggested (Binarova et al., 2006; Pastuglia et al., 2006), but the molecular mechanisms behind this remain to be uncovered.
γ-Tubulin interacts with the transcription factor E2F1 and modulates progression through the G1/S phase of the cell cycle in mammalian cells (Hoog et al., 2011). In Arabidopsis thaliana there are three E2Fs: E2FB and E2FA are positive regulators for cell proliferation and endoreduplication, respectively, while E2FC suppresses cell division (Magyar et al., 2016). E2Fs form heterodimers with either dimerization partner A or B (DPA and DPB) proteins in order to bind to target promoters (Magyar et al., 2000). Arabidopsis E2Fs are controlled by the RETINOBLASTOMA-RELATED (RBR) protein, and the E2F binding activity of RBR is regulated through phosphorylation by the CYCLIN-DEPENDENT-KINASE A;1 (CDKA;1)/CYCLIN-Ds (CYCDs)/KIP-RELATED PROTEINs (KRPs). This E2F–RBR system controls the expression of genes involved in cell cycle progression, DNA replication, and chromatin dynamics. The E2F–RBR pathway was primarily linked to the regulation of cell cycle entry during G1/S progression, but its function was also associated with G2/M regulation, as part of the evolutionarily conserved multiprotein DP, RB-like, E2F, and MuvB (DREAM) complex (Magyar et al., 2016). In addition, E2FA, when released from RBR repression, is important for the switch from cell division to endoreduplication (De Veylder et al., 2002; Magyar et al., 2012).
In this work, we decided to characterize whether the microtubule-independent role of plant γ-tubulin in cell division (Binarova et al., 2006) is performed through γ-tubulin interaction with plant E2F transcription factors. Our data suggest that γ-tubulin has a regulatory role together with E2FA, E2FB, or E2FC to coordinate cell division with the endocycle during plant growth and development.
Materials and methods
Vector construction and plant transformation
As described in detail in (Binarova et al., 2006), the γ-tubulin RNAi vector was constructed by generating an inverted hairpin loop of a 722 bp fragment corresponding to nucleotides 700–1425 of A. thaliana TubG1. In brief, the PCR fragment was cloned directly into the pGEM-T vector (Promega) and cloned into pART69 to generate the RNAi intermediate construct and then transferred into an ethanol-inducible vector to generate pGreenAlcA:TubG1-RNAi. Alternatively, a hairpin from a 552 bp fragment corresponding to nucleotides 292–844 of the AtTubG1 sequence was used. The PCR fragments were cloned into pHANNIBAL (Wesley et al., 2001) and the resulting cassette, consisting of the Cauliflower mosaic virus (CaMV) 35S promoter, two hairpin arms separated with an intron, and the OCS terminator, was transferred to the binary vector pART27 using NotI cleavage. Gene-specific primers used for pArt27:TubG1-RNAi construction are listed in Supplementary Table S1 at JXB online. Arabidopsis thaliana (Col-0) was transformed by the floral dip method in a suspension of Agrobacterium tumefaciens GV3101.
To construct the pgE2FB–green fluorescent protein (GFP) and pgE2FC–GFP translational fusions, the promoter regions and the genomic clones including exons and introns were amplified from Col-0 genomic DNA using the primer combinations described in Supplementary Table S1. In both cases, the genomic sequences were fused at the 3' end with the coding sequence of GFP in a pGreenII-based pGII0125 destination vector (Galinha et al., 2007) by using the Invitrogen 3 way gateway system (Invitrogen, USA). Transgenic plants were generated as described before (Henriques et al., 2010).
Plant material and growth conditions
Arabidopsis thaliana ecotype Col-0 wild type (WT), pArt27:TubG1-RNAi, pGreenAlcA:TubG1-RNAi (SEM analyses), the e2fa-2;e2fb-1 double mutant (Heyman et al., 2011), and lines expressing GFP-tagged E2Fs under the control of their own promoters pgE2FA–GFP (Henriques et al., 2010), pgE2FB–GFP, and pgE2FC–GFP were used. Seeds were sterilized and grown vertically on Murashige and Skoog (MS) agar plates with appropriate selection (Kohoutova et al., 2015) under a 16 h:8 h, light:dark regime. For anti-microtubule drug treatment, MS agar medium was supplied with 1 µM amiprophosmethyl (APM; Duchefa Biochemie A0185) from stock solution in DMSO. Plants were observed at the developmental stage of 11 DAS (days after sowing).
Interaction studies with in vitro translated proteins
The coding regions of γ-tubulin, DPA, DPB, E2FA, E2FB, E2FC, and MYB66 were inserted into pEU3-NII-HLICNot and pEU3-NII-GLICNot vectors by ligation-independent cloning. The coding region of γ-tubulin was inserted into a pEU3-NII-HxHLICNot vector (data not shown) encoding a double-hexahistidine (His12) tag to increase the sensitivity of detection (Khan et al., 2006). The truncated E2F mutants were amplified from the WT E2F-containing vectors by using the primers listed in Supplementary Table S2. The vector constructs were used for in vitro translation as described previously (Nagy and Meszaros, 2014).
E2F–γ-tubulin interactions were investigated with pull-down assay by using 1.875 µl of Pierce Glutathione Magnetic Beads (Thermo Scientific) and 30 µl of His6-γ-tubulin and glutathione S-transferase (GST)–E2FA/B/C-containing in vitro co-translation mixtures. Prior to the 1 h incubation with the beads, the translation mixtures were diluted six times with wash buffer (125 mM Tris, 150 mM NaCl, 0.5% Triton-X, pH 8). The specificity of E2F–γ-tubulin interaction was demonstrated by mixing and incubating 20 µl of GST–MYB66/E2FA and 40 µl of His12-γ-tubulin-containing in vitro translation mixtures for 1 h at room temperature and pulling down the complexes with 2.5 µl of Pierce Glutathione Magnetic Beads. Following the pull-down, the beads were split and washed with either 150 mM or 500 mM NaCl containing wash buffer. The DP–γ-tubulin competition experiments were implemented by mixing and incubating 30 µl of His6-DPA/B-containing translation mixture and 1.875 µl of Pierce Glutathione Magnetic Beads pre-coated with E2F–γ-tubulin complexes for 1 h at room temperature. Interaction between the truncated E2FA/B/C proteins and DPB–γ-tubulin was demonstrated by mixing and incubating 15 µl of truncated GST–E2FA/B/C and 15 µl of His6-DPB–γ-tubulin-containing in vitro translation mixtures for 1 h at room temperature and pulling down the complexes with 1.875 µl of Pierce Glutathione Magnetic Beads. The beads were washed with 250 mM NaCl-containing wash buffer.
The samples were subjected to SDS–PAGE and the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane by semi-dry blotting. The membranes were blocked with 5% non-fat milk powder in phosphate-buffered saline (PBS) containing 0.05% Tween-20. The labelled proteins were detected either directly by anti-polyHis-peroxidase (POD) (Sigma) diluted 1:2000 or by rabbit anti-GST (Upstate) diluted 1:2000 and the secondary antibody goat anti-rabbit–horseradish peroxidase (HRP) (Cell Signaling) diluted 1:5000.
dsDNA-binding site studies with ALPHA and EMSA
Complementary oligonucleotides with Cy5 or biotin labelling on the upper strand and 3' inverse dT modification on the lower strand were synthesized to generate the consensus E2F-binding site and its mutant version (IBA Life Sciences). Sequences of WT E2F and mutated binding sites: upper strand 5'-TTTCCCGCCAAtagc-3'; lower strand 5'-gctaTTGGCGGGAAA-3' and 5'-TTTCCaaCCAAtagc-3'; 5'-gctaTTGGttGGAAA-3', respectively. Lower case letters represent overhanging and mutated nucleotides (modified from Magyar et al., 2012). The oligonucleotides were hybridized in annealing buffer (100 mM Tris, 500 mM NaCl, pH 8) to create 50 μM dsDNA stock solutions.
ALPHA (amplified luminescence proximity homogeneous assay) sample mixtures were assembled in a 384-well plate at 20 μl final volume (AlphaPlate 384 SW, Perkin Elmer). The measurements were implemented in PBS-diluted wheatgerm translation mixtures with BSA and salmon sperm DNA added to reach 1 mg ml–1 and 0.1 μg ml–1 final concentrations, respectively. AlphaScreen Streptavidin Donor and Nickel chelate Acceptor beads (Perkin Elmer) were used at 20 μg ml–1 final concentration. Diverse variations of in vitro translated His6-E2FB, His6-DPB, and GST–γ-tubulin (~50 ng) were mixed and incubated at room temperature for 2 h. Following the addition of biotin-labelled dsDNA-coated donor beads, the mixtures were incubated for 30 min. Next, the acceptor beads were added, and the samples were incubated for a further hour. The luminescent signal (counts per second; cps) was measured by using the default ALPHA settings of Enspire (Perkin Elmer) plate reader.
EMSA was carried out by using a 5% native polyacrylamide gel (29:1 acrylamide:bisacrylamide) polymerized in 50 mM Tris, 0.38 M glycine, and 2 mM EDTA, pH 7.5-containing buffer. The His6-E2FB, His6-DPB, and GST–γ-tubulin were produced by wheatgerm in vitro translation. The respective Cy5-labelled dsDNAs were incubated with 1 μl of the relevant total translation mixture at 5 μM final DNA concentration for 1 h at room temperature in EMSA buffer [250 ng of poly(dI–dC), 10 mM Tris, 50 mM KCl, 1 mM DTT, and 1% Tween-20, pH 7.5]. The samples were loaded into a native polyacrylamide gel after mixing them with loading buffer without dyes and separated at 70 V in TBE buffer. The gels were imaged using a Typhoon scanner.
Supershift assays were performed as described by Hsieh et al. (2016) with slight modifications. The GST–E2FA, untagged DPB, and His6-γ-tubulin were produced by wheatgerm in vitro translation. GST–E2FA and His6-γ-tubulin were co-translated, GST–E2FA and untagged DPB were pre-incubated for 90 min, then 1.6 μl of the respective translation mixtures were incubated for a further 90 min prior to addition of DNA. The supershift assay mixtures contained 1 μl of anti-polyHis antibody (Sigma, H1029), 1 μg ml–1 poly(dI–dC), and 0.1 μM Cy5-labelled WT DNA. Of note, the available GST antibodies did not work in the supershift assay; thus, His6-γ-tubulin was used to demonstrate DNA binding of heterotrimer complex. All the in vitro interaction studies were performed in at least five independent experiments.
Protein purification, immunoblotting, and immunoprecipitation
Proliferating or non-proliferating conditions were induced by transfer of Arabidopsis seedlings expressing GFP-tagged E2Fs to MS liquid medium containing 2% or 0% sucrose, respectively, for 6 h. Total protein was extracted from Arabidopsis seedlings as described (Magyar et al., 1997) but protease inhibitor cocktail (Sigma-Aldrich) was used instead of unique protease inhibitors. Immunoprecipitations (IPs) by using GFP–Trap-coupled magnetic beads (ChromoTek) were carried out according to Magyar et al. (2012). Bound proteins were separated by SDS–PAGE and immunoblotting assays were carried out as described (Henriques et al., 2010). Antibodies used in immunoblotting experiments were: anti-γ-tubulin plant-specific antibody AthTU (Drykova et al., 2003), anti-DPA (Magyar et al., 2005), anti-DPB (Umbrasaite et al., 2010), and mouse anti-GFP monoclonal (Roche). Five independent IP experiments were performed, and representative images are shown.
ChIP
For ChIP in seedlings, WT (Col-0) and e2fa-2;e2fb-1 double mutant plants were grown on sucrose-free medium in 12 h light:12 h dark conditions. On the morning of the 12th day, half of the plants of both genotypes were kept in the dark for a further 4 h (extended dark). A 3 g aliquot of the ‘extended dark’ and ‘light’ plants was cross-linked with 1% formaldehyde. For ChIP in cell culture, 1 g of exponentially grown Col-0 cells (3 d after subculturing) were cross-linked with 1% formaldehyde. Cross-linking and chromatin isolation were performed based on Saleh et al. (2008). For sonication of the chromatin, BioRupter (Diagonade) was used with 30 s on/off cycles 10 times at high mode. Chromatin fragments were precipitated using 15 µl of anti-γ-tubulin antibody and collected with protein A–agarose/salmon sperm DNA (Millipore). The purified DNA was used in quantitative PCRs (qPCRs) to amplify promoter regions with specific primers listed in Supplementary Table S1. Relative enrichment was calculated by dividing the antibody IP signals by the no-antibody signals. Three independent experiments were performed.
RNA extraction and qRT-PCR
Total RNA was extracted from whole-plant tissues using an RNeasy Mini Kit (Qiagen), cDNA synthesis was carried using 1 µg of RNA with a Maxima First Strand cDNA synthesis kit with double-stranded DNase (Thermo Scientific). For the qPCRs, Sybr Green JumpStart Taq ReadyMix (Sigma) was used and the reactions were performed using Rotor-Gene Q (Qiagen). We used actin as a reference gene in all qRT-PCR experiments as described in Horvath et al. (2017). Raw data were analysed with the ΔΔCT method. Gene-specific primers used for qRT-PCR analysis are listed in Supplementary Table S1. Three independent experiments were performed.
Phenotype analyses of γ-tubulin RNAi plants
Immunofluorescence labelling and fluorescence microscopy
Slide preparation of squashed Arabidopsis seedlings and immunolabelling were performed as described in Horvath et al. (2017) using anti-α-tubulin (Abcam, 1:1000) and anti-mouse Alexa Fluor 488 (1:600) antibodies. Chromatin was counterstained with DAPI. Images were obtained by the Olympus IX-81 FV-1000 confocal imaging system as decribed in Kohoutova et al. (2015). Images were analysexd and prepared using FV10 ASW2.0 (Olympus) and Adobe Photoshop/Illustrator CS4 (Adobe System), respectively. Cell counting was performed on immunolabelled mesophyll cells of the WT and γ-tubulin RNAi leaves. Cells were counted manually in randomly selected areas defined by a 1000 µm×1000 µm square using ImageJ software.
5-Ethynyl-2'-deoxyuridine (EdU) labelling
EdU labelling of whole mount seedlings was performed using the Click-iT EdU Alexa Fluor 488 HCS Assay (Molecular Probes) according to Horvath et al. (2017). Seedlings were incubated in liquid medium with a EdU pulse at a dilution of 1:1000 for 1 h.
Flow cytometry
Nuclei were isolated from WT control (Col-0) and γ-tubulin RNAi seedlings as described in Doskocilova et al. (2013) using the Partec CyStain UV precise P kit (Partec), and flow cytometry was measured on a BD LSRII (BD Biosciences) with a solid state laser (excitation 405 nm) and 450/50 band pass filter. Data evaluation was performed in FlowJo from at least six independent experiments. The endoreduplication index (EI) was determined from percentage values of each C-level with the formula: EI=[(0×% 2C)+(1×% 4C)+(2×% 8C)+(3×% 16C)+(4×% 32C)]/100 (Radziejwoski et al., 2011). Chromocentres were labelled and counted as described previously by Doskocilova et al. (2013). Phenotype analyses were performed from at least six independent experiments, and representative images are shown.
TEM
Root tips of 6-day-old A. thaliana expressing GFP-tagged E2FA under its own promoter were high-pressure frozen in Leica EM PACT2 using 200 mM sucrose in 10 mM Tris buffer, pH 6.6, as a cryoprotectant. Frozen samples were transferred under liquid nitrogen to an automatic freeze-substitution machine (Leica EM AFS2 equipped with Leica EM FSP) and freeze-substitution was done using dehydrated acetone containing 0.2% uranyl acetate, 0.2% glutaraldehyde, and 5% water for 24 h with a gradual temperature increase to –80 °C, 24 h at –80 °C, and increased to –50 °C during the following 30 h. The mixture was replaced with pure dehydrated acetone and kept for 6 h at –50 °C followed by –45 °C for 4 h. Samples were then infiltrated with Lowicryl HM20 (Polysciences, Inc.) resin at –45 °C: 33% mixture with acetone for 12 h, 66% resin in acetone for 12 h, and three replacements of pure resin for 12, 12, and 11 h. The resin was polymerized using UV light, starting at –45 °C with a gradual temperature increase to –35 °C during 24 h, increased to 20 °C during 1 h, and the polymerization continued for the next 72 h.
Sections (70–90 nm) were cut using aLeica EM UC6 ultramicrotome with a diamond knife (Diatome AG, Biel, Switzerland) and mounted on gilded copper slots with a formvar film. To block unspecific immunogold labelling, slots were pre-incubated with 10% normal goat serum (Invitrogen), 0.2% cold water fish skin gelatin, and 1% BSA in PBS with 0.1% Tween-20, pH 7.4 (PBTB) for 20 mins. The sections were incubated with chicken anti-GFP antibody diluted 1:100 and rabbit anti-γ-tubulin [plant-specific antibody (Drykova et al., 2003) or DQ-19, Sigma-Aldrich] in PBTB containing 0.2% cold water fish skin gelatin for 1 h, washed in 0.005% Tween-20 in PBS, pH 7.4 (PBT), and incubated for 1 h with 6 nm gold-conjugated donkey anti-chicken IgG and 12 nm gold-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA), each diluted 1:30 in PBTB, and 0.2% cold water fish skin gelatin. Control incubations were performed without primary antibody. The slots were observed in an FEI Morgagni 268 transmission electron microscope at 80 kV equipped with a Mega View III CCD camera (Olympus) or in an FEI Tecnai 20 G2 electron microscope equipped with a Gatan 863 Tridiem Imaging Filterand Model 894 UltraScan 1000 camera.
SEM
Arabidopsis leaves were fixed with 3% (w/v) glutaraldehyde in cacodylate buffer, pH 7.2, and washed in the same buffer. Then the samples were dehydrated in an alcohol series (25%, 50%, 75%, 90%, 96%, and 100%) followed by absolute acetone, and dried in a critical-point device (Balzers 010, Balzers Union Ltd, Lichtenstein). The dried samples were mounted onto aluminium stubs, sputter-coated with gold (Polaron SC 510, Watford, UK), and examined on an Aquasem (Tescan, Brno, Czech Republic) scanning electron microscope at 15 kV in high vacuum mode.
Accession numbers
TubG1, At3g61650; E2FA, At2g36010.3; E2FB, At5g2220.2; E2FC, At1g47870.1; DPA, At5g02470.1; DPB, At5g03415.1; MYB66, At5g14750.1; CDKB1;1, At3g54180; CYCD3;1, At4g34160; CCS52A2, At4g11920; PCNA1, At1g07370; RBR, At3g12280; ORC2, At2g37560.
Results
γ-Tubulin binds to E2FA, E2FB, and E2FC proteins in vitro
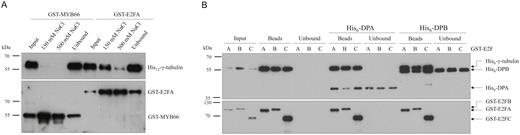
Our previous finding on the nuclear localization of γ-tubulin and the meristem organization phenotypes of γ-tubulin-silenced plants (Binarova et al., 2000, 2006) implied a cell cycle-related function for γ-tubulin. This is consistent with reports on animal cells, where γ-tubulin associates with E2F1 to regulate cell proliferation (Hoog et al., 2011). To test whether γ-tubulin specifically interacts with E2Fs also in plants, we inserted the protein coding sequences of γ-tubulin, cell division-controlling E2FA, and cell fate-determining MYB66 transcription factors into vectors with appropriate tags that enable the purification and detection of in vitro translated proteins (Supplementary Table S3). The obtained pull-down results demonstrated that γ-tubulin and E2FA form a complex even under stringent washing conditions, while no detectable amount of γ-tubulin is associated with the MYB66-coated magnetic beads (Fig. 1A).
γ-Tubulin interacts specifically with E2FA, E2FB, and E2FC in vitro and their interaction is not affected by DPs. (A) In vitro translated His12-γ-tubulin was incubated with GST-tagged E2FA and MYB66 transcription factor, glutathione-coated magnetic beads were added to the mixture, and the non-specifically bound proteins were removed by washing with either 150 mM or 500 mM NaCl. (B) The GST-tagged E2FA, E2FB, and E2FC proteins were co-translated with His6-γ-tubulin and the complexes were isolated by glutathione-coated magnetic beads. The pulled-down complexes were complemented with either His6-DPA- or His6-DPB-containing translation mixtures (marked above the blots) to study if the DP disrupts the E2F–γ-tubulin interaction. (A, B) Input, bead-bound and unbound proteins were transferred to a PVDF membrane and probed with anti-polyHis-POD (upper parts) and anti-GST (lower parts).
Next, we co-translated hexaHis-labelled γ-tubulin and GST-tagged E2FA, E2FB, and E2FC proteins for pull-down experiments, and showed that all three E2Fs have the ability to interact with γ-tubulin (Fig. 1B). To determine whether the presence of E2F DPs disrupts the γ-tubulin–E2F interaction, we added DPA- and DPB-containing wheatgerm translation extract to the co-translated complexes of E2FA, E2FB, or E2FC with γ-tubulin and subsequently isolated the E2F protein complexes by glutathione-coated magnetic beads. As shown in Fig. 1B, the amount of γ-tubulin in complex with E2Fs was not reduced after addition of DPs and the DPs simultaneously with γ-tubulin associated with E2Fs. Furthermore, the detection of DP proteins in the unbound fraction of pull-down experiments suggests that even a surplus of DPs cannot disrupt the already formed γ-tubulin–E2F complexes (Fig. 1B). Together our data suggest that binding of γ-tubulin and DP to E2Fs is independent and the proteins may form a heterotrimeric complex.
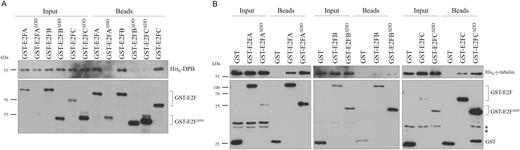
To test whether the dimerization domain (DD) on the E2Fs is required for the γ-tubulin interaction, we prepared constructs to in vitro translated C-terminally truncated E2Fs up to the DNA-binding domain. Indeed these E2Fs without the DDs: E2FAΔDD, E2FBΔDD, and E2FCΔDD, lost the ability to associate with DPB (Fig. 2A). However, all three truncated E2Fs could still bind to γ-tubulin in pull-down assays, suggesting that plant E2Fs do not require the DD for interaction with γ-tubulin (Fig. 2B). These data further support that γ-tubulin and DP are able to simultaneously associate with E2Fs and form a ternary complex.
The E2F dimerization domain (DD) is not essential for γ-tubulin binding. (A) In vitro translated wild-type and truncated (ΔDD) GST-tagged E2FA, E2FB, and E2FC proteins were incubated with His6-DPB and the complexes were isolated by glutathione-coated magnetic beads to demonstrate that His6-DPB does not bind to the truncated variants of E2Fs. (B) In vitro translated GST, wild-type, and truncated (ΔDD) GST-tagged E2FA, E2FB, and E2FC proteins were incubated with His6-γ-tubulin and the complexes were isolated by glutathione-coated magnetic beads to study if the truncated variants of E2Fs can bind His6-γ-tubulin. *Non-specific bands. (A, B) Input and bead-bound proteins were transferred to PVDF membrane and probed with anti-polyHis-POD (upper parts) and anti-GST (lower parts).
γ-Tubulin forms a complex with E2FA, E2FB, and E2FC in vivo
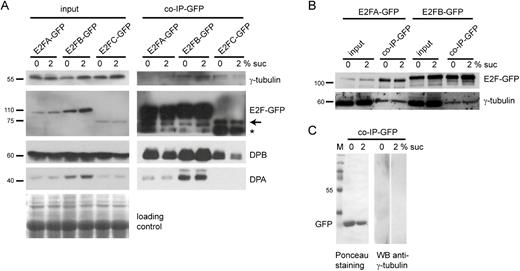
To study whether plant E2Fs may interact with γ-tubulin in vivo, we utilized transgenic Arabidopsis plants expressing GFP-tagged E2FA (pgE2FA–GFP lines 81 and 82), E2FB (pgE2FB–GFP line72), and E2FC (pgE2FC–GFP line 2/8/3) under the control of their respective own promoters. Because sucrose can have an effect on E2F activity, we treated seedlings with 2% sucrose or no sucrose for 6 h (Magyar et al., 2012). We then immunopurified E2FA–GFP, E2FB–GFP, and E2FC–GFP through the GFP tag and tested for the presence of DPA and DPB proteins and γ-tubulin (Fig. 3A). As expected, the E2F–GFP proteins together with the DPs were enriched in the GFP pull-down compared with the crude extract. In agreement with our in vitro data (Figs 1, 2), γ-tubulin was also detectable in association with E2FA–GFP, E2FB–GFP, and E2FC–GFP; however, in contrast to DPs, it was clearly less abundant in the immunoprecipitate than in the crude extract. This suggests that only a minor portion of the total γ-tubulin pool associates with E2FA, E2FB, and E2FC in vivo. The amount of γ-tubulin co-immunoprecipitated with E2Fs did not consistently differ in the presence or absence of sucrose (Fig. 3B), suggesting that the formation of the complex is not affected by conditions that are known to alter RBR phosphorylation and cell proliferation. To rule out that γ-tubulin non-specifically associates with the beads or with the GFP tag, we purified GFP from GFP-expressing plants (35S:GFP) and found no detectable γ-tubulin in the GFP pull-down (Fig. 3C). Our data are in agreement with in vitro results which show that E2FA, B, and C are able to bind the DPs and γ-tubulin simultaneously (Figs 1, 2).
The activator E2FA and E2FB and the repressor E2FC transcription factors interact with γ-tubulin in Arabidopsis seedlings. (A) Proteins were co-immunoprecipitated (Co-IP) from seedlings (7 DAS) expressing E2FA–GFP (line 81), E2FB–GFP (line 72), or E2FC–GFP (line 2/8/3) grown with or without sucrose (2% or 0% Suc, respectively) for 6 h. Input and co-IP protein samples are indicated on the top left and right side, respectively. Western blot detection was performed with antibodies against GFP, γ-tubulin, DPA, and DPB, indicated on the right side. Arrow, E2FC–GFP. Asterisk, non-specific band. Molecular weight standards are indicated on the left side. Loading control: Coomassie-stained proteins on the membrane. (B) Proteins were Co-IP from seedlings (10 DAS) expressing E2FA–GFP (line 82) and E2FB–GFP (line 72) grown with or without sucrose (2% or 0% Suc, respectively) for 6 h. Input and co-IP protein samples are indicated on the top. Western blot detection with antibodies against GFP and γ-tubulin. (C) GFP-expressing seedlings were used as control. Western blot detection with anti-γ-tubulin antibody (right). GFP is demonstrated by Ponceau staining (left). M, molecular weight standards.
γ-Tubulin is localized together with E2FA in the nuclei
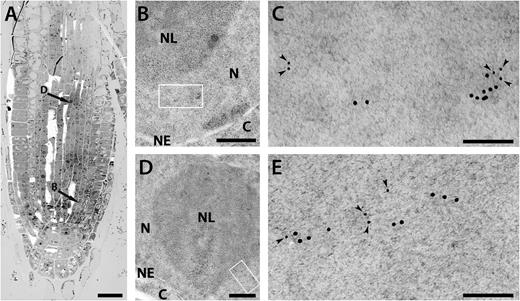
Nuclear localization of plant γ-tubulin was documented by us previously (Binarova et al., 2000; Chumova et al., 2018), and the presence of a nuclear localization sequence (NLS) and nuclear export sequence (NES) in γ-tubulin supports that its nuclear traffic is regulated (Supplementary Fig. S1). It was also shown that E2FA is predominantly a nuclear protein (Magyar et al., 2012). We performed TEM immunogold labelling using anti-γ-tubulin antibody in root tips of plants expressing E2FA–GFP under the native promoter. High-pressure freezing and freeze-substitution resulted in good preservation of the ultrastructure including the nuclei (Fig. 4A, B, D). Multiple clusters of γ-tubulin were immunogold labelled on ultrathin sections in the nucleoplasm and sometimes in the nucleolus. Double-immunogold labelling of γ-tubulin and E2FA–GFP showed that some fraction of γ-tubulin and GFP labelling co-localized (Fig. 4C, E; Supplementary Fig. S2). Two different anti-γ-tubulin antibodies (AthTU and DQ-19) revealed a similarly clustered immunogold labelling pattern that partially co-localized with E2FA–GFP labelling (Fig. 4C, E).
Ultrastructural immunolocalization of γ-tubulin and GFP–E2F in nuclei of A. thaliana root meristem cells. (A) Electron micrograph of a root tip section at low magnification. Arrows indicate the cells shown at higher magnification in images (B) and (D). Scale bar: 20 µm. (B, D) Part of the root cell nucleus double-immunogold-labelled with anti-GFP antibody and anti-γ-tubulin antibody DQ-19 (B) or AthTU (D) showing morphology of the cellular compartments. N, nucleoplasm; NL, nucleolus; C, cytoplasm; NE, nuclear envelope. Scale bar: 500 nm. (C, E) High magnification of the area shown by the rectangle in (B) and (D), respectively. Small gold particles (6 nm, indicated by arrowheads) are E2F–GFP; large gold particles (12 nm) are γ-tubulin (C, DQ-19; E, AthTU). Scale bar: 100 nm.
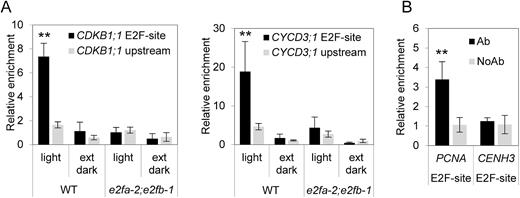
γ-Tubulin binds to promoters of G1/S and G2/M regulators in an E2FA- and E2FB-dependent manner
To investigate whether γ-tubulin is recruited to the promoters of E2F target genes, ChIP was performed with Col-0 plants (Fig. 5A). We used conditions when the growth is maximal, 4 h in the light period, or when growth is arrested by a 4 h extended dark period (Graf et al., 2010). We found enrichment of γ-tubulin binding by ChIP on the promoter region of the G1/S regulator CYCD3;1 and the mitotic CDKB1;1 in the proliferation-promoting light condition but not in the proliferation-repressing dark condition (Fig. 5A). The enrichment was specific to the region of these promoters where the E2F-binding site is located. In the e2fa-2;e2fb-1 double mutant, the γ-tubulin enrichment with the CYCD3;1 and CDKB1;1 promoters was not detectable, suggesting that the E2Fs are required for γ-tubulin recruitment (Fig. 5A). We also tested whether γ-tubulin may associate with the promoter of a typical S-phase-specific E2F target, proliferating cell nuclear antigen (PCNA). For this, we used fully proliferating cultured cells and we found a clear enrichment at the E2F site of the PCNA promoter compared with the no antibody control (Fig. 5B).
γ-Tubulin binds to promoters of E2F-regulated genes. (A) ChIP analysis showed γ-tubulin association with the promoter regions of CDKB1;1 and CycD3 in wild-type (WT) plants grown in proliferative conditions (light) but not in the e2fa-2;e2fb-1 double mutant plants. AthTU antibody was used for immunoprecipitation, and DNA regions containing binding sites for E2F (E2F-site) were analysed. Upstream regions were used as negative controls. (B) ChIP analysis showed γ-tubulin association with promoter regions of the E2F-regulated replication gene PCNA in dividing cultured Col-0 cells, while there was no binding to the CENH3 promotor. No antibody (NoAb) was used as control. Error bars indicate the SD. A non-parametric Mann–Whitney U-test was performed to test for differences between site-specific DNA content in ChIP samples (E2F-site versus upstream region; **P<0.01).
The ChIP results are consistent with our protein interaction data on binding of γ-tubulin to the E2F–DP heterodimer in vitro and in vivo. To investigate whether γ-tubulin alone or as an E2F–DP–γ-tubulin complex may directly bind to DNA with an E2F site, we utilized EMSA by applying in vitro translated proteins and fluorescently labelled DNA either with the consensus E2F-binding site or with the E2F site mutant. As expected, either E2FB or DPB alone could not bind the E2F element, but the E2FB–DPB heterodimer did, as shown by the mobility shift with the DNA containing the intact E2F site. The binding of E2F–DP to DNA was not observed with the mutant E2F element (Supplementary Fig. S3A). γ-Tubulin alone or together with E2FB did not bind to the E2F site, suggesting that γ-tubulin–E2F heterodimers have no ability to bind DNA. The addition of γ-tubulin did not alter the binding of E2FB–DPB heterodimer to the E2F site and the binding was also observed when γ-tubulin was pre-incubated with E2FB to form a heterodimer and DPB was added later (Supplemenrtary Fig. S3A). These data indicated an ability of E2F–DP–γ-tubulin heterotrimer to bind to DNA containing an E2F site. Though we were not able to separate the E2F–DP heterodimer and E2F–DP–γ-tubulin heterotrimer by EMSA, addition of the antibody recognizing the His tag fused with γ-tubulin resulted in a supershift of the E2F–DP–γ-tubulin complex (Fig. S3B).
To gain further quantitative evidence on γ-tubulin DNA binding, we also assayed direct protein–DNA association by ALPHA using biotin-labelled DNA and in vitro translated proteins. The DNA–E2F interaction was detected by the streptavidin donor and Ni chelate acceptor beads, and measuring the luminescence signal. In agreement with the results we obtained by EMSA, only the E2FB–DPB heterodimer produced luminescence above the background level with the E2F site-containing DNA, but not with its mutant form. This proximity-generated fluorescence signal between ther E2FB–DPB heterodimer and the DNA was reduced when γ-tubulin was added (Supplementary Fig. S4). This observation indicates that γ-tubulin may modulate E2F–DP association with DNA through ternary complex formation.
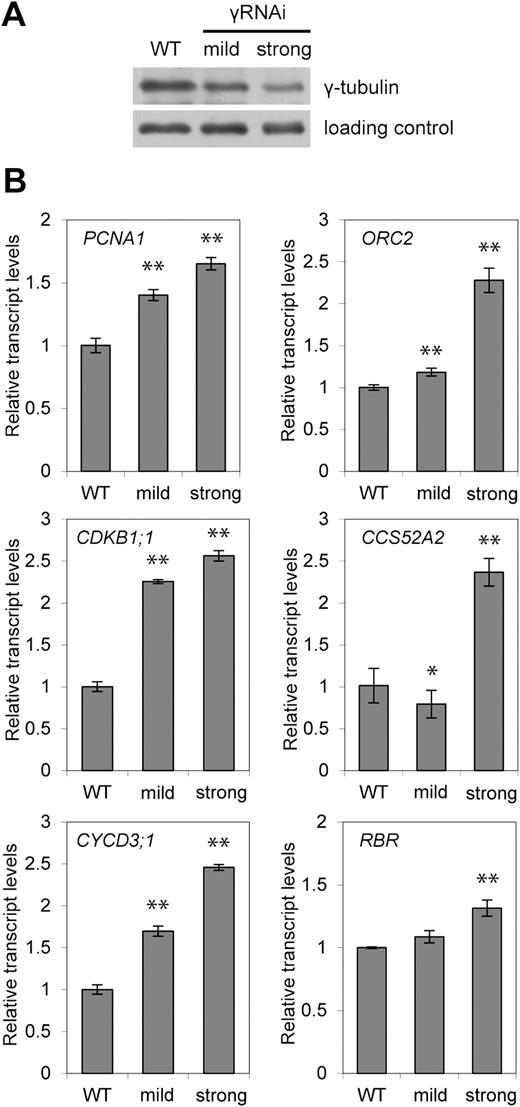
Expression of E2F target genes is elevated in plants with reduced γ-tubulin levels
We have found that γ-tubulin forms protein complexes with E2Fs, co-localizes with E2FA in the nucleus, binds to promoters of E2F target genes in an E2F-dependent manner, and modulates binding of the E2F–DP heterodimer to DNA at the E2F site. To investigate how the expression of selected E2F target genes is affected by γ-tubulin, we analysed the mRNA levels of genes involved in proliferation and the endocycle in seedlings where γ-tubulin was silenced by RNAi (Binarova et al., 2006). qRT-PCR analyses were performed on young newly emerged leaves of the WT control and γ-tubulin RNAi plants with mild and strong silencing effects (Fig. 6A). The expression of the PCNA1, ORC2, CDKB1;1, CCS52A2, CYCD3;1, and RBR genes was found to be elevated in comparison with the WT, and the up-regulation of the expression correlated with the strength of γ-tubulin silencing (Fig. 6B). Our data suggest that γ-tubulin acts as a repressor of E2F-regulated genes at G1/S and G2/M transitions and at the switch to the endocycle.
Expression of E2F target genes is up-regulated proportionally to the strength of the γ-tubulin RNAi silencing. (A) Protein level of γ-tubulin in WT and γ-tubulin RNAi (γRNAi) plants (10 DAS) with mild and strong silencing effects. Proteins in the total extract were probed on Western blot with anti-γ-tubulin antibody. (B) Expression levels of the indicated genes were determined by qRT-PCR from seedlings of WT and γ-tubulin RNAi plants with mild and strong silencing effects (10 DAS). A non-parametric Mann–Whitney U-test was performed to test for differences between the wild type and γ-tubulin RNAi (*P<0.05; **P<0.01). Error bars indicate the SD of values obtained from three independent experiments in triplicate.
Silencing of γ-tubulin leads to ectopic cell divisions and enhanced levels of endoreduplication
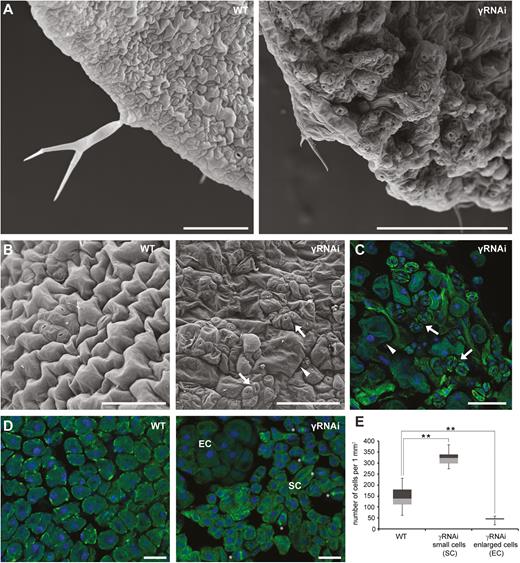
As we published previously, a reduction of γ-tubulin levels affects cell division in both roots and aerial parts of γ-tubulin RNAi Arabidopsis plants (Binarova et al., 2006). We performed SEM and showed that the leaves of γ-tubulin RNAi plants developed a rough blade surface as compared with leaves of the WT control that have lobed pavement cells and regular pattern of stomata (Fig. 7A). More detailed inspection of the epidermis of γ-tubulin RNAi leaves by SEM and by immunofluorescence showed that the cellular outcome of γ-tubulin silencing was mixed, and both enlarged isodiametric cells as well as small cells and stomatal clusters were observed (Fig. 7B, C). In contrast to leaf epidermal cells of RNAi plants (Fig. 7B, C), the WT leaves grown in the presence of the anti-microtubular drug, APM, showed only large swollen pavement cells with no lobes, an effect typically found when microtubules are depolymerized (Supplementary Fig. S5). The consequence of γ-tubulin silencing was also observed in the mesophyll leaf cells. We observed groups of cells with a centrally localized large nucleus and dense cytoplasm that were ~2.5 times smaller than the average cell size in the WT, while there were also cells that were enlarged and appeared vacuolated, a sign that is typical of non-dividing cells (Fig. 7D, E). The population of the small meristematic-like cells showed 4.0% (n=3533) division, while hardly any division was observed in mesophyll cells of the WT control (n=1274; Fig. 7D, E).
Silencing of γ-tubulin leads to ectopic division and stomatal clustering. (A) Representative SEM images of leaves of control WT and γ-tubulin RNAi (γRNAi) Arabidopsis plants (12 DAS). (B) SEM images, abaxial epidermis of the WT and γ-tubulin RNAi (11 DAS) with clusters of meristemoids and stomata (arrows), and enlarged isodiametric cells (arrowhead). Scale bars: 100 µm. (C) Immunofluorescence labelling of α-tubulin (green) in the abaxial epidermis of γ-tubulin RNAi (11 DAS) with a cluster of stomata (arrows) and enlarged isodiametric cells (arrowhead). DNA stained by DAPI (blue). Scale bar: 100 µm. (D) Immunofluorescence labelling of α-tubulin (green) in mesophyll cells in WT and γ-tubulin RNAi leaves (11 DAS) with small proliferating cells (SC, mitosis, and cytokinesis marked by asterisks) and enlarged isodiametric cells (EC); DNA stained by DAPI (blue). Scale bars: 150 µm. (E) Box plot of cell number counted in a randomly selected defined area (1000 µm×1000 µm) of mesophyll cells of WT and γ-tubulin RNAi leaves. Cell numbers are visualized in quartiles of ranked data expressed by means with the SD (n=22). Asterisks indicate P<0.01 comparing WT and γ-tubulin RNAi by Student’s t-test. Compared with the WT with cells of uniform size, two distinct cell populations of small and enlarged cells were demonstrated in γ-tubulin RNAi leaves.
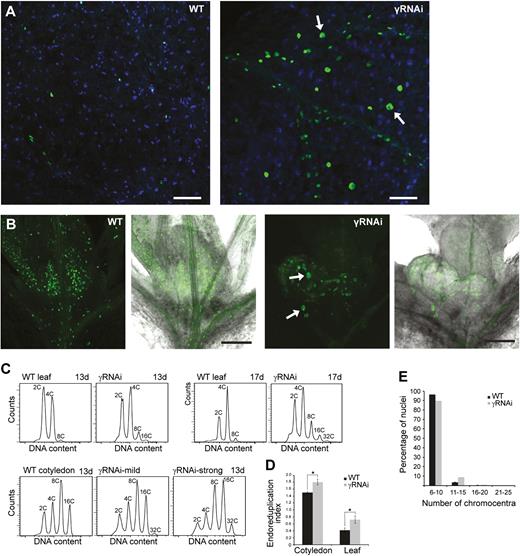
We applied EdU labelling to visualize DNA replication in γ-tubulin RNAi plants (Fig. 8A, B). After a 1 h EdU pulse, large EdU-positive nuclei were observed in cotyledons of RNAi plants, but not in the WT cotyledons (Fig. 8A). As expected, many nuclei with uniform size were labelled with EdU in the youngest leaf of the WT seedlings (10 d) after a 1 h pulse of EdU treatment. However, in leaves where γ-tubulin was silenced, we observed a mixture of large nuclei with EdU signal and EdU-positive nuclei typical of WT leaves at this time point (Fig. 8B). This observation suggests that the enlarged cells with large EdU-positive nuclei in the γ-tubulin RNAi line represent cells that prematurely entered the endocycle. Further support for over-endoreduplication upon γ-tubulin silencing comes from flow cytometry measurements of DNA content. As compared with WT control, the DNA content was elevated in cotyledons as well as in young leaves of RNAi plants (Fig. 8C). The degree of endoreduplication was quantified by the EI (the number of endoreduplication cycles per cell). There was an almost 2-fold increase of the EI in leaves with reduced γ-tubulin levels compared with theWT and a 1.2-fold increase of the EI in cotyledons (Fig. 8D). To prove that the higher DNA content in plants with silenced γ-tubulin is due to endoreduplication and not polyploidy, we counted chromocentres in WT and γ-tubulin RNAi plants. Distribution of cells with the given number of chromocentres showed a distinct peak corresponding to the diploid number of chromosomes, and was similar for WT and γ-tubulin RNAi leaves (Fig. 8E). Thus, the increased ploidy found upon γ-tubulin silencing is not due to endomitosis, but endoreduplication. The increase in the endoreduplication level shown by microscopic and flow cytometry data is consistent with the elevated expression of the CCS52A2 gene upon silencing of γ-tubulin (Fig. 6B).
Elevated level of endoreduplication in plants with silenced γ-tubulin. (A) Representative images of EdU (1 h pulse) labelling in large nuclei (arrows) in cotyledons of γ-tubulin RNAi (γRNAi) plants (7 DAS), but not WT control plants: EdU (green), DAPI (blue). Scale bars: 100 µm. (B) EdU labelling (1 h pulse) in the youngest leaves of the WT and γ-tubulin RNAi plants (10 DAS). Large EdU-positive nuclei (arrows). Scale bars: 100 µm. (C) Representative DNA content histograms determined by flow cytometry of WT and γ-tubulin RNAi leaves (13 and 17 DAS) and cotyledons (13 DAS). (D) Endoreduplication index determined from flow cytometry data measured in leaves and cotyledons of WT and γ-tubulin RNAi. Data are represented as mean with the SD (n=8). An asterisk indicates P<0.05 comparing WT and γ-tubulin RNAi in Student’s t-test. (E) Distribution of nuclei with a given number of chromocentres in γ-tubulin RNAi (n=513) and WT leaves (n=1091).
Altogether, our data showed that γ-tubulin silencing results in overproliferation in some cells and enhanced endoreduplication in others, a phenotype that is reminiscent of when E2FA is overexpressed together with DPA (De Veylder et al., 2002; Magyar et al., 2012).
Discussion
The presence of γ-tubulin in plant and animal nuclei suggests non-canonical roles in nuclear processes (Binarova et al., 2000; Chumova et al., 2018; reviewed in Chumova et al., 2019). Here we provide several lines of evidence that plant γ-tubulin has functions in the regulation of cell cycle genes in association with E2F transcription factors: (i) γ-tubulin is in the nucleus and co-localizes with E2F transcription factors; (ii) all canonical Arabidopsis E2F transcription factors, E2FA, E2FB, and E2FC, form complexes with γ-tubulin both in vitro and in vivo, and the formation of the complex is independent of the association of these E2Fs with DPs; (iii) γ-tubulin is able to associate, as part of the E2F–DP complex, with the DNA at the E2F site in vitro and it is recruited in an E2F-dependent manner to the promoters of E2F target genes involved in cell cycle regulation in vivo; (iv) the expression of cell cycle E2F target genes, including the G1/S regulatory PCNA, ORC2, and CYCD3;1 and the G2/M phase-specific CDKB1;1, is derepressed in plants with a silenced level of γ-tubulin; and (v) the observed cellular phenotypes, such as ectopic cell divisions in some cells and extra endoreduplication in others, are in line with functions described for E2Fs.
In contrast to what was shown in animal cells (Hoog et al., 2011), we provide evidence that in plants γ-tubulin can interact not only with the activator-type E2Fs, E2FA and E2FB, but also with the repressor type E2FC. Moreover, we find that the DD is not required for these interactions and, in agreement, γ-tubulin does not compete with DPs to form an alternative E2F–γ-tubulin heterodimeric complex, but it associates with the E2F–DP dimer to form an E2F–DP–γ-tubulin heterotrimeric complex. This ternary interaction of γ-tubulin with E2F–DP heterodimers might influence the DNA binding activities of the E2F–DP complexes and/or the formation of repressor complexes. This is consistent with our ChIP data showing that the γ-tubulin association with chromatin is E2F dependent (Fig. 5) and also with our data on the repressive function of γ-tubulin in the regulation of E2F target gene expression (Fig. 6).
In mammalian cells, γ-tubulin was shown to interact with E2F1–E2F3 to repress their activities in order to regulate the G1/S transition following centrosome duplication. Specifically, the repression of CycE expression by γ-tubulin was suggested to have a role in preventing the re-duplication of centrosomes (Hoog et al., 2011). In higher plant cells, there are no centrosomes, but the dispersed microtubule nucleation sites containing the γ-tubulin complex are also known to be coordinated via the core cell cycle regulators (Binarova et al., 1998; Weingartner et al., 2001). Here we show that besides the role in microtubule nucleation, γ-tubulin also has a function to form repressor complexes together with the canonical E2F transcription factors and thereby regulate the G1/S control point. Thus this function of γ-tubulin appears to be evolutionarily conserved between animals and plants. However, our finding that γ-tubulin together with E2Fs also regulates the G2/M transition and the endocycle is novel and might be plant specific. Accordingly, while γ-tubulin was only detected in G1 and S phase nuclei of mammalian cells (Hoog et al., 2011), plant γ-tubulin was detected in nuclei during G1, S, and G2 phases of the cell cycle (Binarova et al., 2000). Hence the plant γ-tubulin might have a broader role throughout cell cycle progression to coordinate microtubule nucleation with cell cycle progression than that which was suggested for animal cells. Maintaining cell division competence is connected with the inhibition of differentiation and the entry into a modified cell cycle with repeated S phases, called the endocycle (Polyn et al., 2015). It was shown that a repressor complex formed around E2FA has an important role in this process (Magyar et al., 2012). Here we present evidence that γ-tubulin, perhaps specifically in plant cells, is also involved in preventing the onset of the endocycle.
Plants in which γ-tubulin is silenced show ectopic divisions in some cells but enhanced endoreduplication levels in others in the shoot, a phenotype that was reported for lines when E2FA and DPA were ectopically overexpressed together (De Veylder et al., 2002; Magyar et al., 2012). These two distinct outcomes of γ-tubulin silencing were proposed to depend on the presence or absence of a mitosis-inducing factor in different cell types (De Veylder et al., 2002). It was shown that CDKB1;1 is a key E2F target that determines the switch from proliferation to the endocycle, and its promoter is controlled by the balance of activator- versus repressor-type E2Fs (Boudolf et al., 2004). In this work. we found that γ-tubulin binds in an E2F-dependent manner to the CDKB1;1 promoter and inhibits its expression. An elevated CDKB1;1 level corroborated with the derepression of other E2F targets, such as CycD3;1, should lead to cell division; however, we find that in some cells there is an extra level of endocycle and cell enlargement. We show that γ-tubulin also represses another E2F target, the CCS52A gene, that is involved in endocycle onset, which could explain the different outcomes, while there are DNA replication genes under γ-tubulin control—ORC2 and PCNA1—that participate both in the normal cell cycle and the endocycle.
CCS52A belongs to the Cdh1 protein family, an activator subunit of the APC involved in the destruction of mitotic cyclins that is required for the switch from mitosis to endoreduplication (Larson-Rabin et al., 2009; Vanstraelen et al., 2009). The E2FA–RBR complex was shown to repress the expression of CCS52As in mitotically active cells to inhibit the entry into endoreduplication (Magyar et al., 2012), and thus the association of γ-tubulin with E2FA to form a repressor complex could explain the extra level of endocycling when γ-tubulin is silenced. Interestingly, γ-tubulin is implicated in the regulation of the APC complex in Aspergillus, probably acting through Cdh1. The failure of APC inactivation in γ-tubulin mutants prevents mitosis (Edgerton-Morgan and Oakley, 2012; Edgerton et al., 2015).
Other emerging data also underline how cytoskeleton structures can regulate nuclear functions in multiple ways, specifically chromatin domain organization, DNA repair, and gene expression (Schrank and Gautier, 2019). γ-Tubulin belongs to the tubulin family of proteins and it was demonstrated to have the ability to form oligomers and polymers in both plant and animal cells (Chumova et al., 2018; Lindstrom and Alvarado-Kristensson, 2018). Nuclear and perinuclear γ-tubulin was suggested to have scaffolding and sequestration roles for the nuclear envelope and chromatin organization, and in regulation of repair and transcription (reviewed in Chumova et al., 2019).
In summary, our findings suggest that the coordination of cell cycle gene expression with the spatial and temporal functions of γ-tubulin in microtubular cytoskeleton organization in mitotic and differentiated cells may have an important role in plant development.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers.
Table S2. List of primers used for the construction of truncated E2F proteins.
Table S3. List of in vitro translated proteins.
Fig. S1. Protein sequence of Arabidopsis γ-tubulin (At3g61650) with the nuclear localization signal (NLS) and nuclear export signal (NES) marked.
Fig. S2. Ultrastructural immunolocalization of γ-tubulin and GFP–EF2 in nuclei of A. thaliana root meristem cells.
Fig. S3. Analysis of the DNA binding capacity of E2F, DP, and γ-tubulin by EMSA.
Fig. S4. Analysis of the DNA binding capacity of different combinations of E2F, DP, and γ-tubulin by ALPHA.
Fig. S5. The effect of the antimicrotubular drug APM on leaf pavement cells of Col-0 Arabidopsis plant.
Acknowledgements
This work was supported by grant P501 15-11657S from Grant Agency of the Czech Republic to PB and by the Institutional Research Concept RVO 61388971. The research was partially supported by Programme for research and mobility support of young researchers from Czech Academy of Sciences MSM200201901 to HK, by the Biological and Biotechnology Research Council BB/M025047/1 to LB, by grant EFOP-3.6.3-VEKOP-16-2017-00009 to BMK, and grant OTKA-NN-111085 (Hungary) to TM, by the grant GINOP-2.3.2-15-2016-00001 from the Ministry of National Economy (Hungary) to ZM, and by the Czech-BioImaging RI project LM2015062 MEYS CR and OP RDE CZ.02. 1.01/0.0/0.0/16_013/0001775 to PH. We thank Barbora Gallová and Anna Kuchařová for help with cloning and selection of γ-tubulin RNAi plants, Gabriela Kočárová for technical assistance, and the CSIRO Plant Industry, Australia for vector pHANNIBAL (Wesley et al., 2001).
Authors contributions
BK, HK, and JC planned and performed experiments, analysed and interpreted the data, and contributed to the manuscript preparation; BK and HK contributed equally. CP, LT, OK, PH, ZM, and VF performed experiments and analysed the data; PB, LB, ZM, and TM discussed, planned, and designed the experiments, and discussed the data; PB wrote the manuscript that was completed and finalized together with LB, ZM, and TM.
References
Author notes
These authors contributed equally to this work.




Comments