-
PDF
- Split View
-
Views
-
Cite
Cite
Barbora Valiskova, Sona Gregorova, Diana Lustyk, Petr Šimeček, Petr Jansa, Jiří Forejt, Genic and chromosomal components of Prdm9-driven hybrid male sterility in mice (Mus musculus), Genetics, Volume 222, Issue 1, September 2022, iyac116, https://doi.org/10.1093/genetics/iyac116
Close - Share Icon Share
Abstract
Hybrid sterility contributes to speciation by preventing gene flow between related taxa. Prdm9, the first and only hybrid male sterility gene known in vertebrates, predetermines the sites of recombination between homologous chromosomes and their synapsis in early meiotic prophase. The asymmetric binding of PRDM9 to heterosubspecific homologs of Mus musculus musculus × Mus musculus domesticus F1 hybrids and increase of PRDM9-independent DNA double-strand break hotspots results indificult- to- repair double-strand breaks, incomplete synapsis of homologous chromosomes, and meiotic arrest at the first meiotic prophase. Here, we show that Prdm9 behaves as a major hybrid male sterility gene in mice outside the Mus musculus musculus × Mus musculus domesticus F1 hybrids, in the genomes composed of Mus musculus castaneus and Mus musculus musculus chromosomes segregating on the Mus musculus domesticus background. The Prdm9cst/dom2 (castaneus/domesticus) allelic combination secures meiotic synapsis, testes weight, and sperm count within physiological limits, while the Prdm9msc1/dom2 (musculus/domesticus) males show a range of fertility impairment. Out of 5 quantitative trait loci contributing to the Prdm9msc1/dom2-related infertility, 4 control either meiotic synapsis or fertility phenotypes and 1 controls both, synapsis, and fertility. Whole-genome genotyping of individual chromosomes showed preferential involvement of nonrecombinant musculus chromosomes in asynapsis in accordance with the chromosomal character of hybrid male sterility. Moreover, we show that the overall asynapsis rate can be estimated solely from the genotype of individual males by scoring the effect of nonrecombinant musculus chromosomes. Prdm9-controlled hybrid male sterility represents an example of genetic architecture of hybrid male sterility consisting of genic and chromosomal components.
Introduction
Hybrid male sterility (HMS), one of the postzygotic reproductive isolation mechanisms, prevents gene flow between related species and enables speciation. Infertility of interspecific hybrids has attracted human attention for a long time; sterility of the mule was already discussed by Aristotle and Darwin (Darwin 1859; Coyne and Orr 2004; Maheshwari and Barbash 2011). Although HMS is the most studied mechanism of reproductive isolation, surprisingly modest progress has been made since the time of Darwin in understanding its genetic architecture. The generally accepted model, known as Dobzhansky–Muller incompatibility (DMI), is based on epistatic incompatibility of independently evolving interacting genes, which results in aberrant interaction of their alleles that have not been tested by natural selection (Dobzhansky 1936; Muller and Pontecorvo 1942; Dobzhansky 1951). Admittedly, not a single case of a pair of HMS genes involved in DMI has been documented until now. HMS known to operate in organisms as distant as plants, animals or fungi shares 2 basic rules suggestive of a common genetic mechanism. Haldane’s rule posits that if in the F1 offspring of sexually reproducing species 1 sex is absent, rare, or sterile, it is the heterogametic (heterozygous) sex (Haldane 1922). Although Haldane's rule was published hundred years ago, the molecular mechanism is still unclear. Another common rule refers to a disproportionately large role of chromosome X (Chr X) compared to that of autosomes, known as the large X-effect or Coyne’s rule (Presgraves 2008). In addition to focusing on identification of HMS genes, the idea of chromosomal HMS had been discussed (Dobzhansky 1951; White 1969) but later refuted as an unlikely primary mechanism (Coyne and Orr 2004).
Most of our current knowledge about hybrid sterility in animals comes from Drosophila studies (Presgraves 2003; Coyne and Orr 2004; Maheshwari and Barbash 2011), but more recently, house mouse subspecies Mus musculus musculus, Mus musculus domesticus, and Mus musculus castaneus (abbreviated here musculus, domesticus and castaneus) proved to be an excellent vertebrate model. During their separation from a common ancestor, domesticus separated first, followed by sister species musculus and castaneus about 130,000–500,000 years ago (Geraldes et al. 2008; White et al. 2009; Duvaux et al. 2011; Suzuki and Aplin 2012; Phifer-Rixey et al. 2020; Fujiwara et al. 2022). Domesticus and musculus formed a narrow hybrid zone across Europe at their secondary contact (Wang et al. 2011; Baird and Macholan 2012; Janoušek et al. 2012), while reproductive isolation between castaneus and domesticus has been weaker (Orth et al. 1998; White, Stubbings, et al. 2012). Somewhat less well defined hybrid zone between musculus and castaneus was reported to follow the Yangtze River in China (Din et al. 1996; Jing et al. 2014), and in Japan, Mus musculus molossinus proved to be a hybrid subspecies with a mosaic genome of musculus (∼95%) and castaneus (∼5%) origin (Abe et al. 2004; Terashima et al. 2006; Yang et al. 2011). The incomplete reproductive isolation was confirmed in laboratory crosses between wild-derived mouse strains representing the 3 subspecies. Fertility phenotypes of musculus × domesticus F1 hybrids showed variations from complete meiotic arrest with small testes and no sperm in the epididymis to apparently normal fertility (Forejt and Ivanyi 1974; Good et al. 2008; Vyskocilova et al. 2009; White et al. 2011; Mukaj et al. 2020; Widmayer et al. 2020), while (castaneus × musculus) and (castaneus × domesticus) F1 hybrids were more fertile, showing partial meiotic arrest and some abnormalities in spermiogenesis, but never full F1 HMS (White, Stubbings, et al. 2012). In accordance with the Large X rule (Coyne 2018), introgressions of musculusPWD Chr X into the domesticusB6 genome and molossinusMSM1 Chr X into the domesticusB6 or domesticusPGN genome by repeated backcrosses resulted in full male sterility (Storchová et al. 2004; Oka et al. 2004, 2010).
In crosses of the musculus- and domesticus-derived inbred strains, we discovered the only known vertebrate HMS gene, Prdm9 (Bhattacharyya et al. 2013, 2014; Gergelits et al. 2019; Forejt et al. 2021). The Prdm9 gene, originally mapped as Hybrid sterility 1 (Hst1) locus (Forejt and Ivanyi 1974), was identified by combination of high-resolution genetic and physical mapping and transgenesis (Gregorová et al. 1996; Trachtulec, Hamvas, et al. 1997; Trachtulec et al. 2008; Mihola et al. 2009; Forejt et al. 2021). The genetic architecture of Prdm9-driven HMS consists in epistatic interaction of 3 major components, Prdm9msc1/dom2 interallelic incompatibility, a modifying effect of the X-linked Hstx2 genetic locus, and musculus/domesticus F1 autosomal heterozygosity. The major phenotypes of Prdm9-driven HMS include small testes without sperm, meiosis arrested at the first meiotic prophase with disturbed synapsis of homologous chromosomes, persisting unrepaired DNA double-strand breaks (DSBs) and impaired male sex chromosome inactivation (MSCI) (Dzur-Gejdosova et al. 2012; Bhattacharyya et al. 2013; Balcova et al. 2016; Gregorova et al. 2018; Forejt et al. 2021). At the molecular level, autosomal asynapsis and male sterility are associated with asymmetric evolutionary erosion of PRDM9 recombination hotspots in musculus and domesticus genomes (Baker et al. 2015; Davies et al. 2016), and with increased occurrence of the PRDM9-independent, default DNA DSB hotspots (Smagulova et al. 2016).
In contrast to the relatively straightforward genetic architecture of Prdm9-driven HMS, the genetic analyses of intersubspecific backcrosses and F2 crosses and studies of wild mice from the European hybrid zone suggested a multigenic or even polygenic control (Good et al. 2008; White, Stubbings, et al. 2012; Turner et al. 2014; Larson et al. 2018). The apparent differences in complexity of genetic control can be partly explained by the fact that different forms of HMS may have different mechanisms and phenotypes, starting from early meiotic arrest in F1 hybrids to mild deterioration of the sperm morphology in multi-generation backcrosses or F2 crosses. The complexity can be further increased by the occurrence of recessive epistatic incompatibilities in the intersubspecific backcrosses and F2 crosses that are silent in F1 hybrids. Other mechanisms of HMS genetic control may be revealed, while some apparent genic complexities may be of chromosomal origin, as discussed herein.
Most of the genetic studies of Prdm9-driven HMS have been performed using only 2 inbred strains, PWD and B6, leaving the possibility that their significance could be limited by their unique genomic makeup. The argument was recently refuted by testing the Prdm9msc1/dom2 genotype on the genetic backgrounds of 16 musculus and domesticus strain F1 hybrids (Mukaj et al. 2020). Here we tested penetrance of the same Prdm9msc1/dom2 allelic combination outside the musculus/domesticus background using a 3-parent hybrid test cross of the musculus, domesticus, and castaneus subspecies. For the first time, we compared observed and estimated asynapsis rate and fertility phenotypes in the same individuals and identified 5 quantitative trait loci that modify the Prdm9msc1/dom2-dependent infertility by controlling meiotic chromosome synapsis or spermatogenic arrest or both.
Materials and methods
Mice and ethics statement
The mice were maintained at the Institute of Molecular Genetics in Vestec, Czech Republic. The CAST/EiJ strain (CAST) was kindly provided by Dr Simon Myers from Oxford University, UK, in 2015. The PWD/Ph strain (PWD) is a wild-derived mouse strain from a single pair of Mus musculus musculus mice trapped in Central Bohemia, Czech Republic, in 1972 (Gregorova and Forejt 2000). The C57BL/6J (B6) inbred strain originates from The Jackson Laboratory, Bar Harbor, ME, USA. All mice were maintained in the Specific Pathogen-Free Facilities, in accordance with animal care protocols approved by the Committee on the Ethics of Animal Experiments of the Institute (No. 41/2012). The animal care obeyed the Czech Republic Act for Experimental Work with Animals (Decree No. 207/2004 Sb and Acts Nos. 246/92 Sb and 77/2004 Sb), fully compatible with the corresponding regulations and standards of the European Union (Council Directive 86/609/EEC and Appendix A of the Council of Europe Convention ETS123).
Genotyping and estimation of chromosome subspecies ancestry
Prdm9msc1, Prdm9cst, and Prdm9dom2 alleles were genotyped using SSLP PCR assay as described previously, with M634F (5′-AACCACCCGGCGATTGA-3′) and M634R (5′-AACCACCCGGCGATTGA-3′) primers producing 215-bp-long amplicon for the B6 allele (chr17: 15,272,538–15,272,752; GRCm38) and approximately 230- and 210-bp-long amplicons for the PWD allele and the CAST allele, respectively. The Hstx2 locus was genotyped with primers SR51F (5′-CAGGAGAAGATGGCACAATA-3′) and SR51R (5′-TAACCCTTTCACCATGTTTC-3′), and the PCR product length of the CAST and PWD alleles was 150 and 130 bp, respectively (ChrX : 67,841,601–67,841,743; GRCm38).
The TC1 males with Prdm9msc1/dom2 genotype were further genotyped by subspecies-specific markers within MiniMUGA array (Morgan et al. 2015; Sigmon et al. 2020). The sex of each sample was confirmed by comparing hybridization intensities for X- and Y-linked markers.
Fertility phenotypes
Males were euthanized by cervical dislocation at 8–12 weeks of age. The body weight (BW) in grams and fertility parameters were determined, weight of paired testes in milligrams (TW) and sperm count in millions. Sperm counting was performed as previously described (Lustyk et al. 2019). Log10(1 + x) (x = sperm count in millions), denoted as log(SC), was applied toward normal distribution and used for QTL analysis. Fertility phenotypes were determined in 310 males of (musculusPWD × castaneusCAST) × domesticusB6 testcross.
Immunostaining and asynapsis rate determination
For immunocytochemistry, spread spermatocyte nuclei were prepared as described (Anderson et al. 1999) with modifications. Briefly, a single-cell suspension of spermatogenic cells in 0.1M sucrose hypotonic solution with protease inhibitors (Roche) was incubated on ice for 10 min and dropped on microscopy slides covered with freshly prepared fixative—1% paraformaldehyde (adjusted pH 8.2 with 0.1 N sodium tetraborate) and allowed to settle for 3 h in a humidified box at 4°C. After brief washing in distilled water and PBS and blocking with 5% goat sera in PBS (vol/vol), the nuclei were immunolabeled using a standard protocol with the following antibodies: anti-HORMAD2 (1:700 dilution, “ab211”) rabbit polyclonal antibody, a gift from Attila Toth) and SYCP3 (1:100, mouse monoclonal antibody, Santa Cruz, # 74569). Anti-γH2AX (1:1,000, rabbit polyclonal, Abcam, ab2893) was used to identify early DSBs and unsynapsed parts of autosomes and sex chromosomes. Secondary antibodies were used at 1:300 dilutions and incubated at 4°C for 60 min: goat anti-Mouse IgG-AlexaFluor568 (MolecularProbes, A-11031), goat anti-Rabbit IgG-AlexaFluor647 (MolecularProbes, A-21245), goat anti-Human IgG-AlexaFluor647 (MolecularProbes, A-21445), and goat anti-Rabbit IgG-AlexaFluor488 (MolecularProbes, A-11034). The images were acquired and examined using a Nikon Eclipse 400 microscope with a motorized stage control using a Plan Fluor objective, 60× (MRH00601; Nikon) and captured using a DS-QiMc monochrome CCD camera (Nikon) and the NIS-Elements program (Nikon). The images were processed using the ImageJ software (Schneider et al. 2012). For each sample we analyzed between 68 and 100 pachynemas. The number of asynapses was scored in each pachynema nucleus. One asynapsis was equal to 1 HORMAD2-stained element, excluding XY chromosomes. The asynapsis rate of each male was calculated as the ratio/percentage of pachynemas with at least 1 asynapsis out of the total number of checked pachynemas. Altogether 120 males of (musculusPWD × castaneusCAST) × domesticusB6 testcross were analyzed for the presence of asynapsed autosomes.
Fluorescent in situ Hybridization (FISH)
Analysis of meiotic synapsis of individual chromosomes was performed by DNA—Fluorescence In Situ Hybridization (FISH) using the XMP XCyting Mouse Chromosome N Whole Painting Probes (Metasystems) as described (Turner et al. 2005) with slight modifications. Testes from 8- to 12-week-old mice were dissected and spread meiocyte nuclei were prepared as described previously (Gregorova et al. 2018) with a modification, which relies on standard immunofluorescence staining followed by DNA FISH. Briefly, after washing and postfixation steps, the slides were dehydrated, denatured and hybridized according to the manufacturer’s recommendations with the XMP Orange XCyting Mouse Chromosome Paint probes for chromosomes 17, 18, and 19: Nos D-1417-050-OR, D-1418-050-OR, and D-1419-050-OR (https://metasystems-probes.com/). After hybridization, the slides were washed in stringency wash solution, drained and mounted in Vectashield DAPI+ mounting medium (Gregorova et al. 2018). The immunostained and FISH painted spread spermatocytes were examined and photographed using a Nikon Eclipse 400 microscope with a motorized stage control using a Plan Fluor objective, 60 (MRH00601; Nikon) and captured using a DS-QiMcmonochrome CCD camera (Nikon) and the NIS-Elements program (Nikon). The images were processed using the ImageJ software (Schneider et al. 2012). Between 68 and 121 pachynemas were analyzed per 1 male. Altogether 32 TC1 males were studied.
QTL mapping
Statistical environment R 3.6.1 and its package “qtl” (Broman and Sen 2009) has been used to perform QTL analysis. MiniMUGA samples data were joined with GigaMUGA samples on shared markers. Only informative SNPs with <10% missing information were selected. Markers’ position and additional annotation were taken from the ‘mini_uwisc_v2.csv’ table obtained from K. Broman’s GitHub repository (https://github.com/kbroman/MUGAarrays). Genotype probabilities between markers were calculated on a grid size of 1 cM or smaller and with genotyping error rate of 1%. Standard interval mapping was performed using scanone function with the Haley-Knott method. Genome-wide significance was calculated by 10,000 permutations and compared to α = 5% threshold. The code can be found at https://github.com/simecek/pwd_cast_b6_tc. QTL mapping of fertility traits and asynapsis was based on 143 and 77 (musculusPWD × castaneusCAST) × domesticusB6 Prdm9msc1/dom2 testcross males, respectively.
Statistics
The statistical significance of differences of the testes weights and sperm counts was assessed by 2-tailed Mann–Whitney test. The asynapsis rate was evaluated by unpaired t-test in the GraphPad Prism version 9.3.1. for MacOS.
Results
Prdm9 controls infertility of male hybrids of 3 mouse subspecies
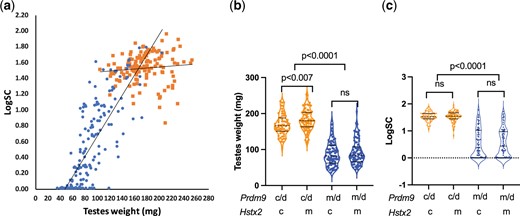
To verify the role of Prdm9 outside musculus and domesticus subspecies, we compared the fertility phenotypes of male hybrids carrying the PWD or CAST allele of Prdm9 from the ♀(musculusPWD × castaneusCAST) × ♂domesticusB6 testcross 1 (hereafter TC1). We estimated testes weight and the number of sperm in the epididymis as quantitative traits in 310 males aged 70–90 days using absolute, rather than relative testes weight values, because the correlation between the testes weight and body weight was not significant in TC1 males (Pearson r = 0.1351, P [2-tailed] 0.1717). The males were genotyped for Prdm9msc1 and Prdm9cst alleles of musculusPWD and castaneusCAST origin and for Hstx2PWD and Hstx2CAST alleles. The males with the Prdm9cst/dom2 genotype were always fertile with 176.0 ± 29.9 mg mean testes weight ± SD and median log-transformed sperm count (log10(SC × 106+1), hereafter log(SC)) 1,543, while the Prdm9msc1/dom2 genotype resulted in continuously varied fertility parameters, from full sterility to full fertility (90.3 ± 33.4 mg mean testes weight ± SD and median log(SC) 0.395 (Fig. 1a and Supplementary Table 1). Thus, the Prdm9 gene acts as a major HMS gene on the mixed genetic background of 3 mouse subspecies. The variable fertility of Prdm9msc1/dom2 TC1 males contrasted with invariable sterility (average testes weight 66.5 ± 1.9 mg, 0.01 × 106 sperm cells) of (musculusPWD × domesticusB6) F1 hybrid males with the same Prdm9msc1/dom2 allelic combination (Supplementary Table 2; see also Supplementary Table 2 in Dzur-Gejdosova et al. [2012]). The difference between TC1 males and (musculusPWD × domesticusB6)F1 indicated involvement of castaneus-derived modifiers in the former cross.
Prdm9 gene and Hstx2 locus control fertility of (musculusPWD × castaneusCAST) × domesticusB6 TC1 males. a) The Prdm9cst/dom2 males are all fertile, while the Prdm9msc1/dom2 allelic combination determines variable fertility phenotypes ranging from complete sterility to fertility. This is in contrast to (musculusPWD × domesticusB6)F1 hybrids where the same Prdm9msc1/dom2 allelic combination causes complete male sterility (insert). b) The Hstx2CAST allele moderately reduces weight of the testes in fertile Prdm9cst/dom2 hybrid males, but c) it does not significantly change the number of sperm in the epididymis. Prdm9 c, d, and m refers to cst, dom2, and msc1 alleles of castaneus, domesticus and musculus origin. Hstx2 c and m refers to cst and msc1 alleles of castaneus and musculus origin.
Next, we focused on the possible involvement of the X-linked HMS locus Hstx2 in fertility variation (Storchová et al. 2004; Bhattacharyya et al. 2014; Lustyk et al. 2019). The only difference ascribed to Hstx2 was a small reduction of the testes weight in fertile Prdm9cst/dom2 males carrying the Hstx2CAST allele (168.5 ± 29.3 mg) compared to Hstx2PWD (181.7 ± 29.0 mg, P = 0.0094, unpaired t-test, Fig. 1, b and c and Supplementary Table 1). This apparently physiological variation did not influence the HMS phenotypes.
We can conclude that the Prdm9 gene acts as a major HMS gene even with admixture of the castaneus genome in the intersubspecific testcross, and the CAST allele of the Hstx2 locus does not differ from the Hstx2PWD allele in controlling the HMS phenotypes of Prdm9msc1/dom2 TC1 males.
QTL mapping of HMS genes segregating in Prdm9msc1/dom2 TC1 males
The variable fertility of the Prdm9msc1/dom2 TC1 males can be caused by reduced DNA DSB asymmetry (Davies et al. 2016; Gregorova et al. 2018) of autosomes that improves their synapsis at meiotic prophase I and/or by the action of modifiers of castaneus origin. To find infertility-modifying genes, we conducted a genome-wide quantitative trait loci (QTL) mapping (Broman and Sen 2009) of male fertility phenotypes using GigaMUGA and MiniMUGA mouse universal genotyping arrays (Morgan et al. 2015; Sigmon et al. 2020). Because each locus in the TC1 cross segregates the PWD and CAST alleles, a new sterility-determining QTL associated with the PWD allele may represent a hitherto unrecognized underdominant F1 HMS locus, while the CAST allele with the same effect can indicate a recessive HMS locus or a castaneus-specific underdominant locus.
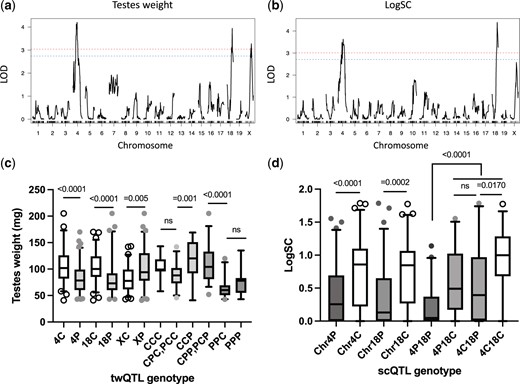
QTL analysis of the panel of 143 Prdm9msc1/dom2 TC1 males revealed 2 autosomal and 1 X-linked locus controlling the weight of testes (twQTL) and 2 autosomal for sperm count (scQTL). The autosomal QTLs for testes weight and sperm count were situated on Chr 4 and Chr 18 (Fig. 2, a and b and Table 1) and their PWD alleles were associated with significantly lower testes weight and sperm count (Mann–Whitney 2-tailed test P < 0.0001). The epistatic effect of Chr 4 and Chr 18 QTL loci on testes weight and sperm count was additive, reducing the size of the testes and the number of sperm with the increasing number of PWD alleles (Mann–Whitney tests P < 0.0001). Unexpectedly, the PWD allele of the X-linked QTL situated distal to Hstx2 locus (138.01 Mb, 103.44 Mb; 142.55 Mb [95% CI]) increased testes weight. Medians for the PWD and CAST alleles were 94 and 77 mg, respectively. The autosomal twQTLs interacted with twQTL-X in an additive-to-recessive manner, so that the simultaneous presence of PWD alleles in autosomal QTLs and PWD or CAST alleles in twQTL-X reduced the testes weight median to 78.0 and 60.5 mg, respectively (Fig. 2, c and d and Supplementary Table 3).
Standard interval QTL mapping of loci controlling fertility phenotypes in Prdm9msc1/dom2 TC1 hybrids. a) Testes weight and b) sperm count as fertility phenotypes. Genome-wide significance thresholds (α = 0.05 and α = 0.10) are indicated by the red and blue dashed line, respectively. c) The effect of individual QTLs on the testes weight and the combined effect of twQTLs grouped according CAST (C) or PWD (P) in the order of alleles of twQTL-4, twQTL-18, and twQTL-X loci. The CAST allele of twQTL-X reduces weight of the testes in contrast to the X-linked Hstx2CAST in the whole set of TC1 males (see Fig. 1c). d) The scQTLs on Chr 4 and 18 display recessive-to-additive epistatic interaction.
QTLs controlling fertility and meiotic asynapsis of Prdm9msc1/dom2 TC1 males.
| Quantitative trait . | Chr . | LOD score . | Positiona . | 95% CI_left . | 95% CI_right . | Cast_mean . | Cast_SEM . | PWD_mean . | PWD_SEM . |
|---|---|---|---|---|---|---|---|---|---|
| Testes weight (mg) | 4 | 4.200 | 83,387,573 | 54,114,217 | 118,968,524 | 107.32 | 4.54 | 83.27 | 3.12 |
| Testes weight (mg) | 18 | 3.942 | 78,293,669 | 68,800,620 | 83,993,897 | 104.77 | 3.44 | 81.78 | 3.98 |
| Testes weight (mg) | X | 3.277 | 137,013,310 | 103,441,512 | 142,549,379 | 82.12 | 3.02 | 103.24 | 4.27 |
| Sperm count (×106) | 4 | 3.629 | 105,294,443 | 54,970,379 | 132,895,458 | 10.68 | 1.8 | 3.8 | 0.79 |
| Sperm count (×106) | 18 | 4.390 | 78,747,720 | 74,832,872 | 88,640,520 | 9.55 | 1.41 | 4.19 | 1.24 |
| Asynapsis rate (%) | 3 | 3.215 | 134,478,458 | 128,224,356 | 144,317,639 | 30.31 | 3.01 | 44.97 | 2.55 |
| Asynapsis rate (%) | 18 | 3.088 | 80,357,856 | 63,255,215 | 89,547,166 | 31.7 | 2.25 | 47.06 | 3.22 |
| Asynapsis rate (%) | X | 3.089 | 82,156,724 | 53,275,228 | 95,160,698 | 31.46 | 2.73 | 45.74 | 2.78 |
| Prdm9a | 17 | n.a. | 15,763,341 | n.a. | n.a. | ||||
| Hstx2b | X | 66.51–69.21 Mb |
| Quantitative trait . | Chr . | LOD score . | Positiona . | 95% CI_left . | 95% CI_right . | Cast_mean . | Cast_SEM . | PWD_mean . | PWD_SEM . |
|---|---|---|---|---|---|---|---|---|---|
| Testes weight (mg) | 4 | 4.200 | 83,387,573 | 54,114,217 | 118,968,524 | 107.32 | 4.54 | 83.27 | 3.12 |
| Testes weight (mg) | 18 | 3.942 | 78,293,669 | 68,800,620 | 83,993,897 | 104.77 | 3.44 | 81.78 | 3.98 |
| Testes weight (mg) | X | 3.277 | 137,013,310 | 103,441,512 | 142,549,379 | 82.12 | 3.02 | 103.24 | 4.27 |
| Sperm count (×106) | 4 | 3.629 | 105,294,443 | 54,970,379 | 132,895,458 | 10.68 | 1.8 | 3.8 | 0.79 |
| Sperm count (×106) | 18 | 4.390 | 78,747,720 | 74,832,872 | 88,640,520 | 9.55 | 1.41 | 4.19 | 1.24 |
| Asynapsis rate (%) | 3 | 3.215 | 134,478,458 | 128,224,356 | 144,317,639 | 30.31 | 3.01 | 44.97 | 2.55 |
| Asynapsis rate (%) | 18 | 3.088 | 80,357,856 | 63,255,215 | 89,547,166 | 31.7 | 2.25 | 47.06 | 3.22 |
| Asynapsis rate (%) | X | 3.089 | 82,156,724 | 53,275,228 | 95,160,698 | 31.46 | 2.73 | 45.74 | 2.78 |
| Prdm9a | 17 | n.a. | 15,763,341 | n.a. | n.a. | ||||
| Hstx2b | X | 66.51–69.21 Mb |
Positions of Prdm9 and Hstx2 are included as a reference. n.a., not applicable.
Position in bp (genome assembly GRCm39).
QTLs controlling fertility and meiotic asynapsis of Prdm9msc1/dom2 TC1 males.
| Quantitative trait . | Chr . | LOD score . | Positiona . | 95% CI_left . | 95% CI_right . | Cast_mean . | Cast_SEM . | PWD_mean . | PWD_SEM . |
|---|---|---|---|---|---|---|---|---|---|
| Testes weight (mg) | 4 | 4.200 | 83,387,573 | 54,114,217 | 118,968,524 | 107.32 | 4.54 | 83.27 | 3.12 |
| Testes weight (mg) | 18 | 3.942 | 78,293,669 | 68,800,620 | 83,993,897 | 104.77 | 3.44 | 81.78 | 3.98 |
| Testes weight (mg) | X | 3.277 | 137,013,310 | 103,441,512 | 142,549,379 | 82.12 | 3.02 | 103.24 | 4.27 |
| Sperm count (×106) | 4 | 3.629 | 105,294,443 | 54,970,379 | 132,895,458 | 10.68 | 1.8 | 3.8 | 0.79 |
| Sperm count (×106) | 18 | 4.390 | 78,747,720 | 74,832,872 | 88,640,520 | 9.55 | 1.41 | 4.19 | 1.24 |
| Asynapsis rate (%) | 3 | 3.215 | 134,478,458 | 128,224,356 | 144,317,639 | 30.31 | 3.01 | 44.97 | 2.55 |
| Asynapsis rate (%) | 18 | 3.088 | 80,357,856 | 63,255,215 | 89,547,166 | 31.7 | 2.25 | 47.06 | 3.22 |
| Asynapsis rate (%) | X | 3.089 | 82,156,724 | 53,275,228 | 95,160,698 | 31.46 | 2.73 | 45.74 | 2.78 |
| Prdm9a | 17 | n.a. | 15,763,341 | n.a. | n.a. | ||||
| Hstx2b | X | 66.51–69.21 Mb |
| Quantitative trait . | Chr . | LOD score . | Positiona . | 95% CI_left . | 95% CI_right . | Cast_mean . | Cast_SEM . | PWD_mean . | PWD_SEM . |
|---|---|---|---|---|---|---|---|---|---|
| Testes weight (mg) | 4 | 4.200 | 83,387,573 | 54,114,217 | 118,968,524 | 107.32 | 4.54 | 83.27 | 3.12 |
| Testes weight (mg) | 18 | 3.942 | 78,293,669 | 68,800,620 | 83,993,897 | 104.77 | 3.44 | 81.78 | 3.98 |
| Testes weight (mg) | X | 3.277 | 137,013,310 | 103,441,512 | 142,549,379 | 82.12 | 3.02 | 103.24 | 4.27 |
| Sperm count (×106) | 4 | 3.629 | 105,294,443 | 54,970,379 | 132,895,458 | 10.68 | 1.8 | 3.8 | 0.79 |
| Sperm count (×106) | 18 | 4.390 | 78,747,720 | 74,832,872 | 88,640,520 | 9.55 | 1.41 | 4.19 | 1.24 |
| Asynapsis rate (%) | 3 | 3.215 | 134,478,458 | 128,224,356 | 144,317,639 | 30.31 | 3.01 | 44.97 | 2.55 |
| Asynapsis rate (%) | 18 | 3.088 | 80,357,856 | 63,255,215 | 89,547,166 | 31.7 | 2.25 | 47.06 | 3.22 |
| Asynapsis rate (%) | X | 3.089 | 82,156,724 | 53,275,228 | 95,160,698 | 31.46 | 2.73 | 45.74 | 2.78 |
| Prdm9a | 17 | n.a. | 15,763,341 | n.a. | n.a. | ||||
| Hstx2b | X | 66.51–69.21 Mb |
Positions of Prdm9 and Hstx2 are included as a reference. n.a., not applicable.
Position in bp (genome assembly GRCm39).
Transmission ratio distortion of twQTL-4
The PWD allele of twQTL-4, represented by gUNC7630359 SNP (Chr 4:83.62 Mb) was transmitted from (musculusPWD × castaneusCAST) females in excess of the CAST allele (84/59, binomial test P = 0.044, TRD 58.7). This QTL may overlap the male QTL on the same chromosome at 116 Mb, which showed deficit of the musculusPWK allele (39.5/60.5) in the domesticusWSB × (musculusPWK × musculusCZECH/II) testcross (Larson et al. 2018). The twQTLs on Chr 18 and Chr X obeyed Mendelian transmission, though the proximal Chr 18 showed an excess of the musculusPWD allele in the B6 × (B6 × B6.PWD-Chr 18) backcross (Gregorová et al. 2008).
The Prdm9 gene and the Hstx2 locus control homologous synapsis in TC1 spermatocytes
Completed pairing and synapsis of all homologous autosomes is an important pachytene checkpoint, preventing formation of aneuploid gametes. We have shown that the Prdm9-controlled HMS is associated with high frequency (80–90%) of pachytene spermatocytes showing incomplete homologous synapsis, and that in the consomic domesticusB6-Chr #PWD/B6 × musculusPWD crosses the probability of synapsis failure depends on the subspecies origin of homologs and their physical length (Gregorova et al. 2018). To sum up, the synapsis of meiotic chromosomes in the musculusPWD × domesticusdom2 model of HMS was shown to be controlled in trans by the Prdm9 gene and the Hstx2 locus, and in cis by the degree of inter-subspecific homology of synaptic partners (Bhattacharyya et al. 2013, 2014; Gregorova et al. 2018; Forejt et al. 2021), depending on the number of symmetric DNA DSBs (Davies et al. 2016; Gregorova et al. 2018).
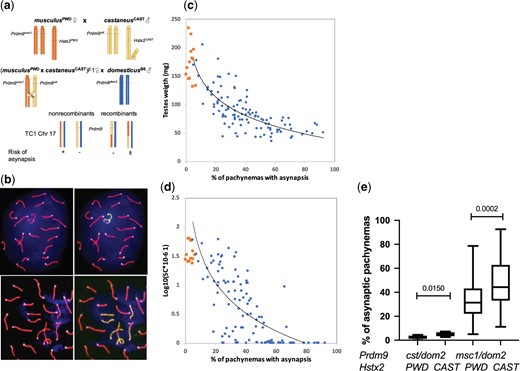
Here we investigated the role of Prdm9 and Hstx2 in meiotic pairing of segregating population of TC1 males. Half of their genome came from the domesticus and the other half from castaneus and musculus subspecies (Fig. 3a). To evaluate homologous synapsis, we visualized the unsynapsed chromosome axes and XY body by immunostaining of SYCP3 and HORMAD2 proteins in pachytene spermatocytes of 108 Prdm9msc1/dom2 males together with 14 randomly chosen Prdm9cst/dom2males (Fig. 3b). The effect of Prdm9 on meiotic chromosome synapsis was dramatic and correlated well with the fertility phenotypes. While the average frequency of pachynemas with 1 or more asynapsed autosomal pair in the Prdm9cst/dom2 males was 3.8% and never exceeded 5%, the average asynapsis rate observed in the Prdm9msc1/dom2 males was 41.4%, ranging from 5% to 85.3%. Moreover, the TC1 males showed an inverse correlation between the testes weight and the frequency of asynaptic pachynemas (Pearson r = −0.6671, P [2-tailed] <0.0001), as well as between the sperm count (log10 transformed) and asynapsis (Pearson r = −0.6911, P [2-tailed] <0.0001) (Fig. 3, c and d and Supplementary Table 4). Surprisingly, the CAST allele of Hstx2 or a closely linked locus, which did not significantly change the testes weight and sperm count, significantly increased the asynapsis rate (Fig. 3e). To summarize, the 25% admixture of the castaneus genome segregating in the TC1 males did not compromise the Prdm9 and Hstx2 control of asynapsis rate. The degree of asynapsis inversely correlated with the fertility phenotypes.
Control of meiotic chromosome synapsis and fertility phenotypes by Prdm9 and Hstx2 genes. a) The subspecific origin of TC1 autosomes is shown on Chr 17 as an example. The probability of homologous synapsis failure mainly depends on the subspecific origin of autosomes and Prdm9 allelic combinations, symbols +, ±, and − refer to high, medium, and low/null probability of asynapsis, respectively. Other factors, such as asyQTLs and MSUC, are not considered here. b) Pachytene spread showing synaptonemal complexes in a fully synapsed cell (upper panel) and a cell with 3 asynapsed pairs of autosomes (lower panel). X–Y chromosomes and asynapsed autosomes are decorated with anti-HORMAD2 antibody (yellow–green). Note weak HORMAD2 staining on the XY chromosomes indicating incomplete MSCI. CEN labels centromeric heterochromatin, and DAPI labels nuclear DNA. c) Relation between asynapsis and testes weight and d) asynapsis and sperm count in Prdm9cst/dom2 (orange) and Prdm9msc1/dom2 (blue) TC1 males. e) The Hstx2CAST allele or a closely linked gene increases asynapsis rate in sterile and fertile TC1 hybrids.
QTL mapping of modifiers of Prdm9-controlled asynapsis
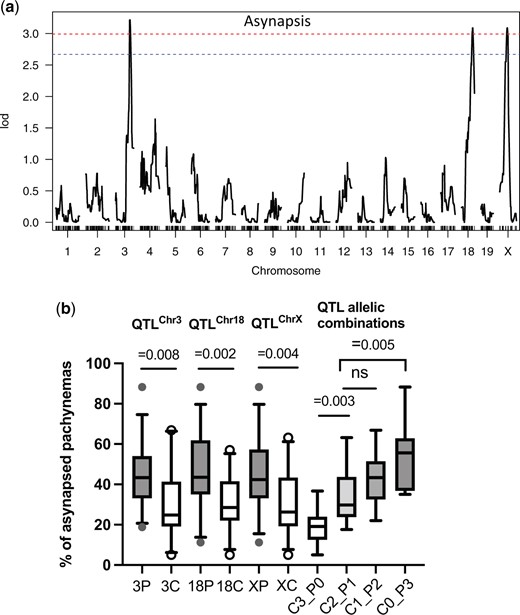
To learn what is the cause of the wide variation of asynapsis rate in Prdm9msc1/dom2 TC1 males, we applied standard interval QTL mapping to 78 Prdm9msc1/dom2 TC1 males for which the asynapsis rate and genotyping data were available. Three QTLs controlling asynapsis rate (asyQTLs) were found, on Chr 3 (134.48 Mb, 128.22 Mb; 144.32 Mb [95% CI]), on Chr 18 (80.36 Mb, 63.26 Mb; 89.55 Mb) and on Chr X (82.16 Mb, 53.28 Mb; 95.16 Mb) (Fig. 4a). While Chr 18 asyQTL (hereafter asyQTL-18) mapped into the same interval as the twQTL-18 for the testes weight, The CAST allele of asyQTL-3 reduced asynapsis of Prdm9msc1/dom2 TC1 males to 30.3% compared to 45.0% of the PWD allele (P = 0.0076 unpaired 2-tailed t-test) without equivalent twQTL or scQTL in the same genomic position. Similarly, the CAST allele of asyQTL-X decreased the average asynapsis to 31.1% compared to 45.7% of the PWD allele (P = 0.0034, t-test) without a corresponding twQTL or scQTL (Fig. 4b and Supplementary Table 5). The additive epistatic interaction between asyQTLs was found with the highest and lowest asynapsis rate associated with 3 (mean 54.1%) and zero (19.1%) PWD alleles of asyQTLs and middle values (34.7% and 43.5%) in the males with 1 or 2 PWD alleles (Fig. 4b). Remarkable is the strong cumulative effect present in the males with CAST alleles in all 3 QTLs, which is independent of the predicted asynapsis rate (see below). There is a caveat when interpreting these QTLs, since the presence of the CAST asyQTL allele on Chr 3 and Chr 18 means that these 2 chromosomes are a priori excluded from the predicted asynapsis, being partially or entirely of the CAST origin.
Meiotic asynapsis as a quantitative trait in Prdm9msc1/dom2 TC1 hybrids. a) Standard interval QTL mapping of loci controlling meiotic chromosome asynapsis. The genome-wide significance threshold (α = 0.05 and α = 0.10) is indicated by the red and blue dashed lines, respectively. b) The asyQTL allelic combinations show additive epistatic interaction between loci on Chrs 3, 18, and X. The average asynapsis rates are shown for subsets of males carrying the CAST allele in all 3, 2, 1, or none asyQTLs (C3_P0, C2_P1, C1_P2, and C_P3). Vertical bars designate 5–95 percentile, and horizontal bars represent medians.
Meiotic homologous synapsis depends on the subspecies origin of homologous chromosomes
Meiotic synapsis in the intersubspecific (musculusPWD × domesticusB6) F1 male hybrids depends on the subspecific origin of homologous chromosomes and is predominantly chromosome-autonomous (Bhattacharyya et al. 2013; Gregorova et al. 2018). The impaired homolog pairing in the intersubspecific F1 hybrids correlates with the asymmetric binding of PRDM9 to heterosubspecific homologs (Davies et al. 2016; Smagulova et al. 2016; Hinch et al. 2019). The asymmetric PRDM9 binding generates asymmetric DNA DSBs that are more difficult-to-repair by homologous recombination and that can deteriorate homologous synapsis (Davies et al. 2016; Gregorova et al. 2018).
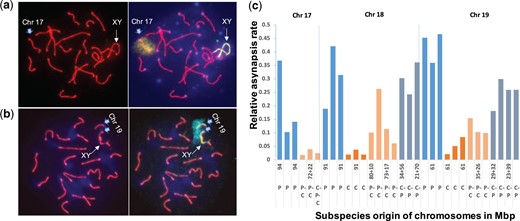
The (castaneusCAST × domesticusB6)F1 males are fertile and show normal homologous synapsis with the mean testes weight 92 mg, sperm count 32.6 × 106 and asynapsis rate 3.2% (Supplementary Table 2; see also White, Ikeda, et al. 2012). We assumed, provided that the genetic background does not abolish the autonomous homolog incompatibility, that the castaneusCAST/domesticusB6 homologs in TC1 pachynemas will pair normally and the musculusPWD/domesticusB6 homologs will be predisposed to asynapsis. To test the assumption, the Chrs 17, 18, and 19 were successively visualized in pachytene spreads by chromosome-specific DNA probes and fluorescence in situ hybridization (FISH) (Fig. 5, a and b). In contrast to uniformly heterozygous (musculusPWD × domesticusB6) F1 hybrids, the TC1 males segregated random stretches of the musculusPWD and castaneusCAST sequence generated by meiotic recombination of their F1 parent (Fig. 3a). For each studied chromosome we selected 3 males with a nonrecombinant musculusPWD and 3 with a nonrecombinant castaneusCAST chromosome, and 3 or 4 males with a recombinant musculus–castaneus or castaneus–musculus (centromere-telomere) chromosome. To compensate for the heterogeneous genetic background, we calculated the relative asynapsis rate of a given chromosome as the ratio between asynapsis rate of that particular chromosome to the overall asynapsis rate. The overall asynapsis rate was defined as a frequency of cells that displayed asynapsis of 1 or more autosomes (Fig. 5c and Supplementary Table 6). The nonrecombinant chromosomes behaved as expected; the relative asynapsis rate of the castaneusCAST/domesticusB6 bivalents did not exceed 10%, while the musculusPWD/domesticusB6 homologs failed to synapse with relative asynapsis rate 10–45%. Unlike the previous study of (musculusPWD × domesticusB6)F1 hybrids (Gregorova et al. 2018), the CAST for PWD sequence substitution in the telomeric ends of recombinant chromosomes improved synapsis more efficiently than an interval of similar length replaced at the centromeric end (Chr 18 P = 0.038, Chr 19 P = 0.010, unpaired t-test).
Relative asynapsis rate of 3 most sensitive autosomes in Prdm9msc1/dom2 TC1 males (see Bhattacharyya et al. 2013; Kauppi et al. 2013; Gregorova et al. 2018). a, b) The average asynapsis rate was estimated by visualization of Chrs 17, 18, and 19 by DNA FISH. a) Fully synapsed Chr 17 is decorated by a Chr 17-specific DNA-painting probe. b) Asynapsed Chr 19 often associates with Chrs X and Y. c) The effect of the CAST sequence on the asynapsis rate of Chrs 17, 18, and 19 in individual TC1 males. The subspecies origin of individual chromosomes coming from a (PWD × CAST)F1 female is either nonrecombinant PWD (P), nonrecombinant CAST (C), recombinant PWD–CAST (P–C, centromere to telomere) or recombinant CAST–PWD (C–P). The length of these chromosomal intervals is given in Mb (genome assembly: GRCm39).
Asynapsis rate predicted from the subspecific ancestry of homologous autosomes
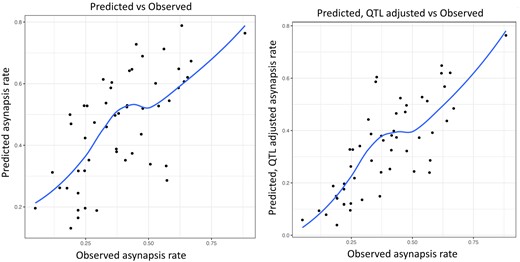
After finding that the cis-acting incompatibility of heterosubspecific homologs operates in a segregating population outside musculusPWD and domesticusB6 crosses, we tried a reverse approach—to predict the overall asynapsis rate solely from the genome-wide genotyping data and to compare it with the observed asynapsis rate. Because the probabilities of asynapsis rates of individual pairs of PWD/B6 homologs in the segregating TC1 males were not known, we arbitrarily used these values from the (musculusPWD × domesticusB6)F1 males [see Fig. 2—source data 1 in Gregorova et al. [2018]). To distinguish the nonrecombinant PWD/B6 pairs of autosomes in the TC1 males, we used >2,000 SNPs of MiniMUGA genotyping arrays to distinguish castaneus from musculus sequence on domesticus background (Morgan et al. 2015; Sigmon et al. 2020). The predicted frequency of pachynemas with 1 or more asynapsed homologs (Supplementary Table 7) was calculated by multiplying probabilities of synapsis of individual autosomes as described previously (Gregorova et al. 2018). Admittedly, there were some caveats limiting this approach. We scored only the nonrecombinant PWD or CAST chromosomes, even though the recombinant PWD chromosomes with less than 27 Mb of the CAST sequence contributed to overall asynapsis rate in the (musculusPWD × domesticusB6)F1 males (Gregorova et al. 2018). Moreover, we had to consider the effect of the newly uncovered asyQTLs, because their PWD/B6 genotype is fixed in (musculusPWD × domesticusB6)F1 males but segregates the CAST and PWD alleles in each asyQTL in individual TC1 males. To bring the predicted asynapsis rates closer to the empirically observed values, we adjusted the predicted asynapsis rates to accommodate the anticipated effect of asyQTLsCAST alleles in each TC1 male. To do so, all Prdm9msc1/dom2 TC1 males were sorted into 4 genotype groups according to the number of CAST alleles of their asyQTLs, namely the groups having 3, 2, 1, or 0 CAST alleles. The group with zero CAST allele (= 3 PWD alleles) mimics the (musculusPWD × domesticusB6)F1 genotype, and was therefore adjusted by coefficient 1.0. In each individual of the remaining groups the predicted asynapsis rates were adjusted by the coefficient defined as a ratio between the mean asynapsis rate of a that genotype group and the mean of the group with 3 PWD alleles (Supplementary Tables 7 and 8). To evaluate the predictions, we analyzed the slope of linear regression of the observed to predicted asynapsis rate and the observed to adjusted, predicted asynapsis rate (Fig. 6). The slopes were significantly different from zero in both cases (predicted asynapsis F = 27.24 DFn, DFd 1.48, P < 0.0001; adjusted, predicted asynapsis F = 52.77, DFn, DFd 1.48, P < 0.0001), confirming the cis-chromosomal mechanism of asynapsis in the Prdm9msc1/dom2 subset of TC1 males. Moreover, the variance of adjusted-predicted to observed asynapsis rate was significantly lower than the variance of predicted to observed asynapsis rate (medians 0.00439 and 0.0132, Mann–Whitney 2-tailed test P = 0.0469), pointing to the role of asyQTLs in the predicted asynapsis rate. To conclude, we found that the asynapsis rate can be predicted based on the subspecies origin of individual chromosomes and on the asyQTLs genotype of each TC1 male.
Spearman correlation between a) predicted and observed asynapsis (Spearman 0.67, RMSE = 0.159) and b) between predicted, QTL-adjusted, and observed asynapsis rate (Spearman 0.77, RMSE = 0.126). The QTL-adjusted, predicted asynapsis rate is significantly closer to the observed asynapsis (Mann–Whitney P = 0.012).
Complex interactions between the observed and predicted asynapsis rate and fertility of Prdm9msc1/dom2 TC1 males
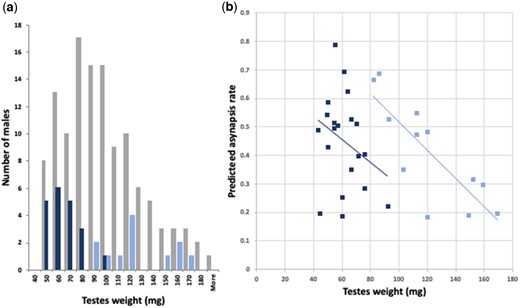
The spermatogenic breakdown of Prdm9msc1/dom2 TC1 males is controlled by chromosomal and genic mechanisms driven by the incompatibility between heterosubspecific homologs and by the QTLs modulating asynapsis rate, testes weight and sperm count. The final phenotype is further affected by downstream consequences, such as asynapsis associated failure of male sex chromosome inactivation (MSCI) or transcriptional silencing of unsynapsed autosomes (MSUC) (Good et al. 2010; Bhattacharyya et al. 2013; Larson et al. 2022). Of 3 asyQTLs that decreased asynapsis rate in the presence of the CAST allele, only asyQTLs-18 reciprocally increased the testes weight (Mann–Whitney 2-tailed test, P = 0.039). The asyQTL-3 and asyQTL-X did not change the fertility phenotypes (P = 0.2104 and 0.8335). Similarly, the CAST alleles of the twQTL-4 and twQTL-X increased or decreased the testes weight, respectively, but without significant effect on the asynapsis rate. The PWD alleles of twQTLs had a particularly strong effect on sterility, overweighing the chromosomal component of meiotic arrest. The most sterile allelic combination (twQTL-4PWD, twQTL-18PWD and twQTL-XCAST) resembled the (musculusPWD × domesticusB6)F1 HMS full meiotic arrest (Forejt et al. 2021), namely the low testes weight (60.9 ± 11.8 mg, mean ± SD) and almost absent sperm in the epididymis (0.30 × 106 ± 0.83, mean ± SD, Fig. 7a). However, on closer look the resemblance was less perfect, because 45% (9/20) of members of this sterile group had the testes weight <60 mg. Such low testes weights do not occur in (musculusPWD × domesticusB6)F1 males and indicate a different mechanism of spermatogenic breakdown dependent on Prdm9msc1/dom2 allelic interaction. Indeed, the correlation was not significant in the sterile group between the predicted asynapsis rate and testes weight, while in the most fertile allelic combination (twQTL-4CAST, twQTL-18CAST, and twQTL-XPWD) the chromosomal component prevailed as shown by significant correlation between the predicted asynapsis rate and testes weight (Fig. 7b).
Distribution of testes weights of Prdm9msc1/dom2 TC1 males with contrasting allelic combinations of twQTLs. a) Gray columns represent all TC1 males tested for twQTL-4, twQTL-18, and twQTL-X. The dark blue columns show testes weights of males with twQTL-4PWD, twQTL-18PWD, and twQTL-XCAST allelic combination (“sterile” allelic combination), while light blue columns refer to twQTL-4CAST, twQTL-18CAST, and twQTL-XPWD males (“fertile” combination). b) The predicted asynapsis rate correlates with testes weight in the “fertile” combination group (blue dots, Spearman r = −0.786, P = 0.0035, 2-tailed), but the correlation is lost in the “sterile” group (orange squares, Spearman r = −0.22, P = 0.34, 2-tailed) indicating the dominance of genic twQTL determinants over chromosomal incompatibility.
Discussion
The long history of genetics of HMS, which began with the seminal papers of Dobzhansky (Dobzhansky 1936, 1951) and Muller (Muller and Pontecorvo 1942), has revealed surprisingly little progress in identification of HMS genes. The current view, mainly based on studies of Drosophila interspecific hybrids, posits HMS as a polygenic trait controlled by mutually interchangeable polygenes with additive effect, or polygenes with complex epistatic interactions. Accordingly, the identified large-effect HMS genes such as OdsH, JYalpha, or Overdrive (Ting et al. 1998; Masly et al. 2006; Phadnis and Orr 2009; Phadnis 2011) are considered unrepresentative exceptions (Presgraves and Meiklejohn 2021). It remains to be seen how general this conclusion is outside Drosophila hybrids. The Prdm9 gene, the first and so far only HMS gene identified in vertebrates, displays some expected and some unorthodox characteristics. Expected for a speciation gene was the rapid evolution and positive selection of the PRDM9 zinc finger DNA-binding domain (Oliver et al. 2009; Buard et al. 2014; Schwartz et al. 2014), but unexpected, considering the DMI hypothesis, was the inter-allelic Prdm9msc1/dom2 incompatibility, shown to be a prerequisite for the Prdm9-driven HMS, as well as substantial intra(sub)specific Prdm9 polymorphism (Grey et al. 2018; Paigen and Petkov 2018; Forejt et al. 2021).
Prdm9 controlled HMS shows high penetrance in the mixed intersubspecific background
The majority of the initial studies, including the positional cloning of Prdm9, were based on a few laboratory inbred strains of domesticus origin and the wild-derived musculus PWD strain (Forejt 1985; Trachtulec et al. 1994; Gregorová et al. 1996; Trachtulec, Mnuková-Fajdelová, et al. 1997; Mihola et al. 2009). The choice of mouse strains was sufficient for genetic studies but could not assess the role of Prdm9 in reproductive isolation between mouse subspecies. The first indication that Prdm9 may operate as a major HMS gene in wild mice came from the crosses of unrelated, wild-derived domesticus and musculus strains (Mukaj et al. 2020). Six out of 7 combinations of Prdm9dom2 or Prdm9dom3 domesticus alleles with Prdm9msc1 or Prdm9msc2 musculus alleles resulted in HMS of F1 hybrid males, regardless of varying wild-derived parts of their genetic background. Moreover, the fertility of hybrids was restored when the ZNF array of Prdm9dom2 allele was replaced with the human PRDM9b allelic form, further pointing to Prdm9 as the major HMS gene (Mukaj et al. 2020). Admittedly, the interrogated allelic combinations were restricted to Prdm9dom2 and various wild-derived musculus alleles and to Prdm9msc1 combined with various wild-derived Prdm9 domesticus alleles in F1 hybrids with invariant Hstx2PWD. More recently, the role of Prdm9 as a major HMS gene was supported by the Prdm9-dependent partial fertility rescue of the fully sterile (Mus spretus × domesticus)F1 interspecific hybrids (Davies et al. 2021). Here we showed the major role of Prdm9 in the fertility and chromosome synapsis of male mice segregating the musculus and castaneus genome on the domesticus background. The Prdm9cst allele ensured physiological values of the testes weight and sperm count, as well as normal course of meiotic pairing, while the Prdm9msc allele of musculus origin induced variable asynapsis rate and meiotic arrest related to the reduced testes weight and sperm count.
Indirect evidence on the role of Prdm9 in fertility of hybrids came from the studies of HMS as a quantitative trait. Underdominant QTLs mapped in backcrosses and F2 crosses to the proximal Chr 17 and Chr X were most likely related to the Prdm9 gene and Hstx2 locus (White et al. 2011; Dzur-Gejdosova et al. 2012; Schwahn et al. 2018). However, as expected, in crosses such as ([domesticusWSB × musculusPWK] × musculusCzechII) (Larson et al. 2018), where Prdm9dom3/msc1 did not segregate, no HMS QTL encompassing the Prdm9 gene was found.
Castaneus modifiers of Prdm9-driven HMS
To map the genetic factors that contribute to variable HMS of Prdm9msc1/dom2 TC1 males, the fertility phenotypes and asynapsis rate were treated as quantitative traits. Of the 5 QTLs found, 2 QTLs controlled the testes weight and sperm count (QTL-4, QTL-X), 2 asynapsis rate (asyQTL-3, asyQTL-X), and, remarkably, only 1 QTL, on Chr 18 (QTL-18) concurrently increased fertility traits and decreased the asynapsis rate. The mechanisms of uncoupling asynapsis rate from testes weight and sperm count are unclear. We can speculate that asyQTLs-3 and asyQTL-X control selective prolongation of pachytene stage for asynaptic pachynemas, without affecting the output of normal spermatocytes. Another explanation could be that a certain part of leptotene/zygotene spermatocytes, which die before developing into asynaptic pachynemas in Prdm9msc1/dom2 TC1 testes, proceed their differentiation to asynaptic pachynemas in (musculusPWD × domesticusB6)F1 hybrid testes (Fig. 8). The tightly X-linked asyQTL-X (X: 82.16 Mb) and Hstx2 (X: 66.5–69.2 Mb) demonstrate the complexity of this control. Both showed only 10% of genetic recombination, yet the CAST allele of Hstx2 increased asynapsis, while the CAST allele of asyQTL-X had the opposite effect in the same set of 70 TC1 males tested. The positive effect of the castaneus allele of twQTL-4 and the negative effect of twQTL-X on testes weight could be explained by a positive and negative effect, respectively, on the length/growth of seminiferous tubuli, influencing the testes weight without changing the proportion of asynaptic pachynemas.
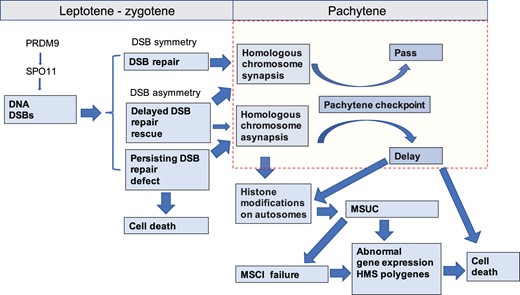
Schematic representation of the events following PRDM9-driven DNA DSB induction during the first meiotic prophase of Prdm9msc1/dom2 intersubspecific male hybrids. The scheme is based on the DNA DSB asymmetry hypothesis (Davies et al. 2016; Gregorova et al. 2018). Increasingly unilateral binding of PRDM9 to its nonself homolog (musculus PRDM9 on domesticus chromosome and vice versa) can increase the risk of delay or failure of DSB repair and immediate cell death before meiotic pairing or can result in pachynemas with incomplete synapsis of homologous chromosomes. Failure to synapse can trigger the pachytene checkpoint and induce meiotic silencing of unsynapsed chromatin (MSUC), resulting in transcriptional silencing in cis of genes essential for meiosis, or in trans by failed transcriptional inactivation of sex chromosomes (MSCI).
An overlapping QTL for the testes weight appeared on Chr 4 in (musculusPWD × domesticusWSB)F2 crosses (White et al. 2011) and in (castaneusCAST × domesticusWSB)F2 crosses (White, Stubbings, et al. 2012). However, a wide range of confidence intervals (98–115 Mb in the former study, 30–118 Mb in the latter and 54–119 Mb in the present study) makes interpretation of their relationship difficult. A possible exception may be a single underdominant QTL on Chr 17 (CI 3–65 Mb) for sperm density as a binary trait in (White et al. 2011), which is indicative of Prdm9msc1/dom2 incompatibility. However, in these and other crosses (Dzur-Gejdosova et al. 2012; Bhattacharyya et al. 2014; Turner and Harr 2014; Wang et al. 2015; Larson et al. 2018; Widmayer et al. 2020), many other QTLs were not shared, supporting the idea of multigenic control of HMS operating distal to the Prdm9/Hstx2-controlled mechanism.
Toward molecular mechanisms of Prdm9-driven HMS genetic architecture
The major causes and consequences of meiotic asynapsis in Prdm9-controlled HMS are associated with DNA DSB repair (Fig. 8). The asymmetric DNA binding of the PRDM9 zinc finger domain to Prdm9 allele-specific binding sites generates asymmetric DNA DSBs that are difficult or impossible to repair (Davies et al. 2016; Smagulova et al. 2016; Hinch et al. 2019). The shortage of symmetric DSBs and increase of PRDM9-independent (default) DSBs (Smagulova et al. 2016) can explain pachytene asynapsis and meiotic arrest in sterile hybrids (Bhattacharyya et al. 2013; Bhattacharyya et al. 2014; Gregorova et al. 2018; Wang et al. 2018; Mukaj et al. 2020). Concurrently, the persisting DSBs can delay cell cycle progression (Hinch et al. 2019) leading either to DSB repair rescue by homologous recombination or to cell death. Even when rescued by noncanonical inter-sister chromatid repair, they can still bring about incomplete synapsis of homologous autosomes and activate the pachytene checkpoint. In addition, the disrupted synapsis can lead to histone modifications and transcriptional silencing of unsynapsed autosomes (Baarends et al. 2005; Turner et al. 2005; Homolka et al. 2007, 2012; Turner 2015) known as Meiotic Silencing of Unsynapsed Chromatin (MSUC) (Schimenti 2005), which can interfere with male sex chromosome inactivation (MSCI) (Forejt 1982, 1984; Turner 2007; Mahadevaiah et al. 2008; Bhattacharyya et al. 2013; de la Fuente et al. 2021; Larson et al. 2022). Inactivation of still-elusive HMS polygenes in asynapsed autosomes by MSUC could provide another explanation to the variable correlation between asynapsis and fertility phenotypes even when the effect of QTLs has been considered. The HMS polygenes silenced by MSUC may be difficult to trace because of their anticipated cumulative and replaceable nature, and because different combinations of autosomes are affected by asynapsis within each TC1 individual.
To conclude, the Prdm9msc1/dom2 allelic incompatibility causes chromosomal HMS of TC1 males by predetermining difficult-to-repair DNA DSBs that preclude full synapsis of homologous autosomes in the first meiotic prophase. The variable HMS penetrance in Prdm9msc1/dom2 TC1 males correlates with the number of nonrecombinant PWD/B6 bivalents and their estimated propensity to asynapsis. This Prdm9-controlled chromosomal HMS is further modified by genes within QTLs controlling testes weight, sperm count and asynapsis rate. We envisage that the Prdm9-controlled HMS may represent a particular case of a hitherto neglected reproductive isolation barrier based on the mutual recognition and synapsis of evolutionary diverging homologous/homeologous chromosomes in meiosis of sexually reproducing species. Experimental models and methodical approaches are available to verify the idea.
Data availability
All mouse strains are available upon request. All raw data and the QTL analysis methods are present within P. Simecek’s GitHub https://github.com/simecek/pwd_cast_b6_tc. The authors affirm that all other data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at GENETICS online.
Acknowledgments
We thank Simon Myers for the B6.Prdm9dom2H mice, Attila Toth for HORMAD2 antibody, and Sarka Takacova for editing the manuscript. We gratefully acknowledge Karel Fusek for discussions and assistance with genotyping.
Funding
This work has been funded by grants from the Czech Science Fundation 20-040 755 and from the LQ1604 Project of the National Sustainability Program II of the Ministry of Education, Youth and Sports of the Czech Republic to J.F.
Conflicts of interest
None declared.