Abstract
Key message
Multiple origins of Indian dwarf wheat were due to two mutations targeting the same TREE domain of a GSK3-like kinase, and these mutations confer to enhanced drought tolerance and increased phosphate and nitrogen accumulation for adaptation to the dry climate of Indian and Pakistan.
Abstract
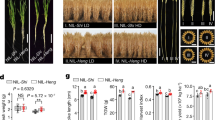
Indian dwarf wheat, featured by the short stature, erect leaves, dense spikes, and small, spherical grains, was a staple crop in India and Pakistan from the Bronze Age until the early 1900s. These morphological features are controlled by a single locus Sphaerococcum 1 (S1), but the genetic identity of the locus and molecular mechanisms underlying the selection of this wheat type are unknown. In this study, we showed that the origin of Indian dwarf wheat was due to two independent missense mutations targeting the conserved TREE domain of a GSK3-like kinase, which is homologous to the Arabidopsis BIN2 protein, a negative regulator in brassinosteroid signaling. The S1 protein is involved in brassinosteroid signaling by physical interaction with the wheat BES1/BZR1 proteins. The dwarf alleles are insensitive to brassinosteroid, upregulates brassinosteroid biosynthetic genes, significantly enhanced drought tolerance, facilitated phosphate accumulation, and increased high molecular weight glutenins. It is the enhanced drought tolerance and accumulation of nitrogen and phosphate that contributed to the adaptation of such a small-grain form of wheat to the dry climate of India and Pakistan. Thus, our research not only identified the genetic events underlying the origin of the Indian dwarf wheat, but also revealed the function of brassinosteroid in the regulation of drought tolerance, phosphate homeostasis, and grain quality.






Similar content being viewed by others
References
Ajmera I, Hodgman TC, Chungui L (2019) An integrative systems perspective on plant phosphate research. Genes (Basel) 20:E139
Albani D, Hammond-Kosack MC, Smith C, Conlan S, Colot V, Holdsworth M, Bevan MW (1997) The wheat transcriptional activator SPA: a seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 9:171–184
Belkhadir Y, Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206:522–540
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Borrell AK, van Oosterom EJ, Mullet JE, George-Jaeggli B, Jordan DR, Klein PE, Hammer GL (2014) Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol 203:817–830
Bu SL, Liu C, Liu N, Zhao JL, Ai LF, Chi H, Li KL, Chien CW, Burlingame AL, Zhang SW, Wang ZY (2017) Immunopurification and mass spectrometry identifies Protein Phosphatase 2A (PP2A) and BIN2/GSK3 as regulators of AKS transcription factors in Arabidopsis. Mol Plant 10(2):345–348
Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, Barajas-Lopez JdD, Fujii H, Wang X (2014) GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci U S A 111:9651–9656
Cheng X, Xin M, Xu R, Chen Z, Cai W, Chai L, Xu H, Jia L, Feng Z, Wang Z, Peng H, Yao Y, Hu Z, Guo W, Ni Z, Sun Q (2020) A single amino acid substitution in STKc_GSK3 kinase conferring semispherical grains and its implications for the origin of Triticum sphaerococcum. Plant Cell 32:923–934
Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol 130:1506–1515
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Feldman M (2001) Origin of cultivated wheat. In: Bojean AP, William JA (eds) The World wheat book–-a history of wheat breeding. Lavoisier Publishing, Paris, France, pp 3–56
Fujihara S, Sasaki H, Aoyagi Y, Sugahara T (2008) Nitrogen-to-protein conversion factors for some cereal products in Japan. J Food Sci 73:C204-209
Gao L, Ma W, Chen J, Wang K, Li J, Wang S, Bekes F, Appels R, Yan Y (2010) Characterization and comparative analysis of wheat high molecular weight glutenin subunits by SDS-PAGE, RP-HPLC, HPCE, and MALDI-TOF-MS. J Agric Food Chem 58:2777–2786
Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S, Sayers L, Doonan JH, Snape JW (2010) A genetic framework for grain size and shape variation in wheat. Plant Cell 22:1046–1056
Gu YQ, Salse J, Coleman-Derr D, Dupin A, Crossman C, Lazo GR, Huo N, Belcram H, Ravel C, Charmet G, Charles M, Anderson OD, Chalhoub B (2006) Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes. Genetics 174:1493–1504
He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A 99:10185–10190
He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307:1634–1638
Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15:2900–2910
Hu Y, Yu D (2014) BRASSINOSTEROID INSENSITIVE2 interacts with abscisic acid insensitive5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26:4394–4408
International Wheat Genome Sequencing Consortium (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Jeong DH, Lee S, Kim SL, Hwang I, An G (2007) Regulation of brassinosteroid responses by phytochrome B in rice. Plant Cell Environ 30:590–599
Kihara H (1944) Discovery of the DD-analyzer, one of the ancestors of Triticum vulgare. Agric Hortic 9:889–890
Kovar JL (2003) Methods of determination of P, K, Ca, Mg and trace elements. In: Peters J (ed) Recommended methods of manure analysis. University of Wisconsin-Extension, Madison, pp 39–47
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295:1299–1301
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222
Li W, Huang L, Gill BS (2008) Recurrent deletions of puroindoline genes at the grain hardness locus in four independent lineages of polyploid wheat. Plant Physiol 146:200–212
Li W, Zhang Q, Wang S, Langham MA, Singh D, Bowden RL, Xu SS (2019) Development and characterization of wheat-sea wheatgrass (Thinopyrum junceiforme) amphiploids for biotic stress resistance and abiotic stress tolerance. Theor Appl Genet 132:163–175
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930
Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24:2168–2183
Liu XY, Lister DL, Zhao Z, Staff RA, Jones P, Zhou L, Pokharia AK, Petrie CA, Pathak A, Lu H, Matuzeviciute MG, Bates J, Lu H, Pilgram TK, Jones MK (2016) The virtues of small grain size: potential pathways to a distinguishing feature of Asian wheats. Quat Int 426:107–119
Long YM, Chao WS, Ma GJ, Xu SS, Qi LL (2017) An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor Appl Genet 130(3):597–607
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
McFadden ES, Sears ER (1946) The origin of Triticum spelta and its free-threshing hexaploid relatives. J Hered 37(81–89):107–116
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Molnár I, Vrána J, Burešová V, Cápal P, Farkas A, Darkó É, Cseh A, Kubaláková M, Molnár-Láng M, Doležel J (2016) Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Plant J 88:452–467
Mori N, Ohta S, Chiba H, Takagi T, Niimi Y, Shinde V, Kajale MD, Osada T (2013) Rediscovery of Indian dwarf wheat (Triticum aestivum L. ssp. sphaerococcum (Perc.) MK.) an ancient crop of the Indian subcontinent. Genet Resour Crop Evol 60:1771–1775
Padmore JM (1990) Protein (crude) in animal feed—dumas method, method No. 968.06. In: Herlrich K (ed) Official methods of analysis of the association of official analytical chemists. AOAC Inc, Arlington, pp 71–72
Percival J (1921) The wheat plant, a monograph. Duckworth and Co, London
Pérez-Pérez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 24:161–173
Peterson CJ, Shelton DR, Martin TJ, Sears RG, Williams E, Graybosch RA (2001) Grain color stability and classification of hard white wheat in the U.S. Euphytica 119:101–107
Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI (2019) Brassinosteroid signaling in plant development and adaptation to stress. Development 146:151894
Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, Leyva A, Rubio V, Sommer H, Paz-Ares J (2014) SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proc Natl Acad Sci U S A 111:14947–14952
Rao MVP (1977) Mapping of the sphaerococcum gene “s” on chromosome 3D of wheat. Cereal Res Commun 5:15–17
Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29:115–125
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Ryu H, Cho H, Bae W, Hwang I (2014) Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun 5:4138
Salamini F, Ozkan H, Brandolini A, Schäfer-Pregl R, Martin W (2002) Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet 3:429–441
Salina E, Borner A, Leonoval I, Korzun V, Laikova L, Maystrenko O, Roder MS (2000) Microsatellite mapping of the induced sphaerococcoid mutation genes in Triticum aestivum. Theor Appl Genet 100:686–689
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak PC, A. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682
Schmidt JW, Weibel DE, Johnson VA (1963) Inheritance of an incompletely dominant character in common wheat simulating Triticum sphaerococcum. Crop Sci 3:261–264
Sears ER (1947) The sphaerococcum gene in wheat. Genetics 32:102–103
Shewry PR, Halford NG, Belton PS, Tatham AS (2002) The structure and properties of gluten: an elastic protein from wheat grain. Philos Trans R Soc Lond B Biol Sci 357:133–142
Singh AP, Fridman Y, Friedlander-Shani L, Tarkowska D, Strnad M, Savaldi-Goldstein S (2014) Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol 166:578–688
Singh AP, Fridman Y, Holland N, Ackerman-Lavert M, Zananiri R, Jaillais Y, Henn A, Savaldi-Goldstein S (2018) Interdependent nutrient availability and steroid hormone signals facilitate root growth plasticity. Dev Cell 46:59–72
Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, Patil S, Kim TW, Ji H, Wong WH, Rhee SY, Wang ZY (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19:765–777
Sun F, Liu X, Wei Q, Liu J, Yang T, Jia L, Wang Y, Yang G, He G (2017) Functional characterization of TaFUSCA3, a B3-superfamily transcription factor gene in the wheat. Front Plant Sci 8:1133
Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39:792–796
Team RC (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Tong H, Chu C (2018) Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci 23:1016–1028
Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24:2561–2577
van Slageren MW (1994) Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Wageningen Agricultural University Papers, Wageningen
Vrána J, Kubaláková M, Šimková H, Cíhalíková J, Lysák MA, Doležel J (2000) Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156:2033–2041
Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol 22:323–325
Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2:505–513
Wang G, Wang G, Zhang X, Wang F, Song R (2012) Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem Anal 23:159–163
Wang J, Luo MC, Chen Z, You FM, Wei Y, Zheng Y, Dvorak J (2013) Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol 198:925–937
Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, Shou H, Mo X, Mao C, Wu P (2014) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci U S A 111:14953–14958
Wang H, Tang J, Liu J, Hu J, Liu J, Chen Y, Cai Z, Wang X (2017) Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol Plant 11:315–325
Wolf A, Watson M, Wolf N (2003) Digestion and dissolution methods for P, K, Ca, Mg and trace elements. In: Peters J (ed) Recommended methods of manure analysis. University of Wisconsin-Extension, Madison, pp 35–36
Yang X, Bai Y, Shang J, Xin R, Tang W (2016) The antagonistic regulation of abscisic acid inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT. Plant Cell Environ 39:1994–2003
Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191
Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38:35–49
Youn JH, Kim TW (2015) Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant 8:552–565
Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, Rodermel S, Yin Y (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65:634–646
Zhang Z, Wang W, Li W (2013) Genetic interactions underlying the biosynthesis and inhibition of beta-diketones in wheat and their impact on glaucousness and cuticle permeability. PLoS ONE 8:e54129
Zhang C, Bai MY, Chong K (2014) Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep 33:683–696
Zhang Z, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li W (2019) Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol J 17:1623–1635
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324
Zhu JY, Li Y, Cao DM, Yang H, Oh E, Bi Y, Zhu S, Wang ZY (2017) The F-box protein KIB1 Mediates Brassinosteroid-Induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol Cell 66:648–657
Zohary D, Hopf M, Weiss E (2012) Domestication of plants in the old world. Oxford University Press, Oxford
Acknowledgements
We thank Dr. Harold Bockelman (USDA ARS, Aberdeen, ID), Dr. Bikram Gill (Kansas State University, Manhattan, KS), Dr. Helmut Knüpffer (The Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), German), Dr. Shuhei Nasuda (Kyoto University, Japan), and Dr. N. Wanatabe (Gifu University, Japan) for providing seeds. We are grateful to Jan Vrána, Zdeňka Dubská, Romana Šperková, and Jitka Weiserová for the technical assistance with chromosome sorting and DNA amplification. This research is supported by the USDA Hatch program through the South Dakota Agricultural Experiment Station and USDA NIFA-IWYP (award number: 2017-67008-25934). IM was supported by the Marie Curie Fellowship grant award “AEGILWHEAT” (H2020-MSCA-IF-2016-746253) and the Hungarian National Research, Development, and Innovation Office (K116277). JD was supported by the ERDF project “Plants as a tool for sustainable global development” (No. CZ.02.1.01/0.0/0.0/16_019/0000827).
Author information
Authors and Affiliations
Contributions
WL conceived the project, all authors conducted experiments and analyzed results, WL drafted the manuscript, and all authors edited and approved it.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Maria von Korff.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, A., Hua, L., Lin, G. et al. Multiple origins of Indian dwarf wheat by mutations targeting the TREE domain of a GSK3-like kinase for drought tolerance, phosphate uptake, and grain quality. Theor Appl Genet 134, 633–645 (2021). https://doi.org/10.1007/s00122-020-03719-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03719-5